Dear All,
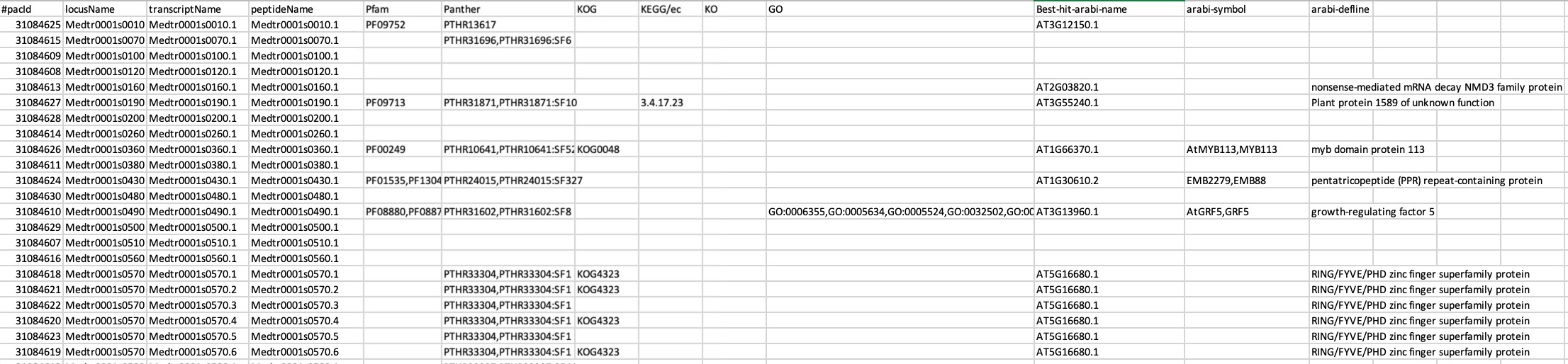
I have a list from gene and annotation file of Medicago truncatula v4 from phytozome. In this example file, they already provide a list of GO-term and kegg pathway. I would like to ask your suggestion about the program that we can use for go-and pathway enrichment with statistical analysis.
Thanks in advance for your help.
Best regards, Yosapol