Entering edit mode
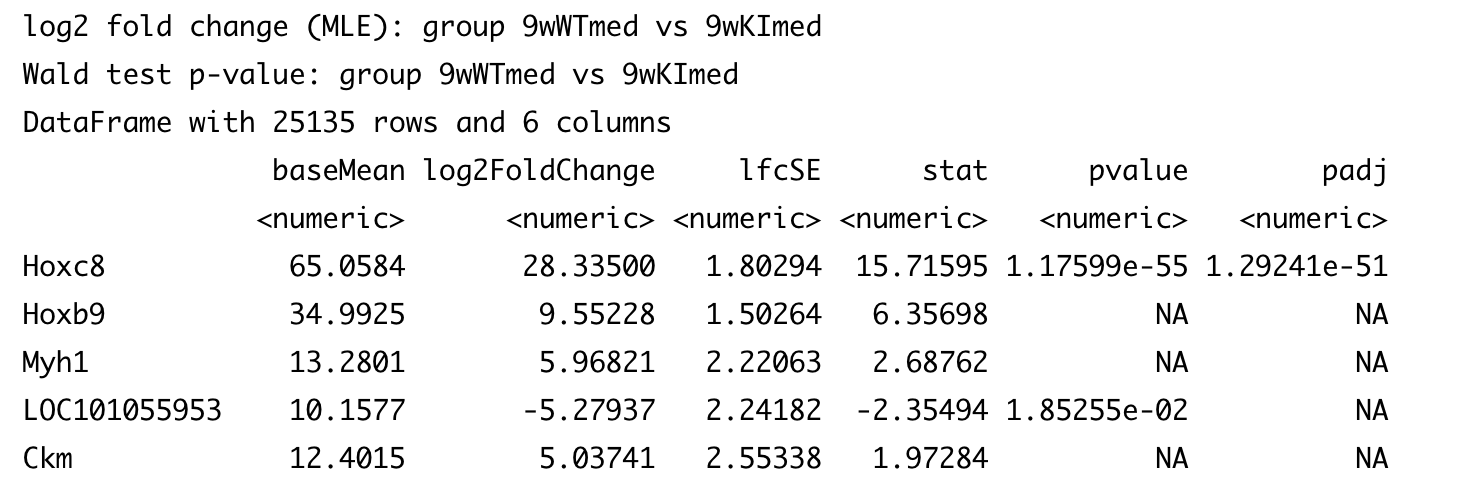
I was trying to identify the genes that are the most different between WT and KI mice based on log 2 fold change. I'm wondering why for some of them it shows NA for p value and p.adj value?
dds <- DESeq(ddsHTSeq_filtered)
# By genotype for med KI vs WT (9w)
res_9w_med_genotype <- results(dds, contrast=c("group","9wWTmed","9wKImed"))
res_9w_med_genotype[order(abs(res_9w_med_genotype$log2FoldChange), decreasing = TRUE),]


http://bioconductor.org/packages/devel/bioc/vignettes/DESeq2/inst/doc/DESeq2.html#why-are-some-p-values-set-to-na
As Mike says, you can be almost certain that most DESeq2 question, unless it is some super special case, is explained in the FAQs.