Entering edit mode
HI,
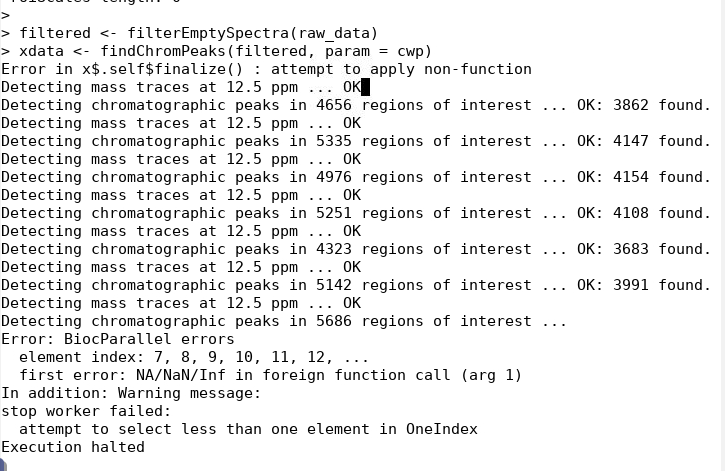
I am trying to run a peakpicking using xcms package and got this error message in one of the modes (didn't have problems with other modes).
could anyone perhaps tell me what has gone wrong and how to fix it? I submitted a batch SLURM job in a closed Linux-based environment using R 4.0.0 so the sessionInfo maybe not as relevant here. Happy to get any tips or advice to try. thanks..
cwp <- CentWaveParam() filtered <- filteredEmptySpectra(raw_data)
``` xdata <- findChromPeaks(filtered, param = cwp)
please also include the results of running the following in an R session
sessionInfo( )