Hi Dear All, When I did RNA-Seq analysis, the GTF file I used was from NCBI. The output of cuffdiff replaced the Gene symbol (official gene symbol) with XLOC's such as:
LOC110534079
LOC110534540
LOC110537830
LOC110485322
LOC110487655
LOC110491675
LOC110492686
LOC110498361
LOC110500236
LOC110502506
Example :
LOC110537830 (ID) = mknk1 (Gene symbol)
Is there any way to convert XLOCs back to Gene symbols?
I searched repeatedly in section Gene in NCBI, but the results Were obtained as LOCs.
I also tried the following sites but it was not successful:
https://www.uniprot.org/uniprot/?query=LOC110525276&sort=score
I also read all the relevant biostars content but did not get the required result: https://www.biostars.org/p/129299/
I also tried the following command in the R software cummeRbund package, but the output answers were as LOCs ID.
cuff <- readCufflinks()
#Retrive significant gene IDs (XLOC) with a pre-specified alpha
diffGeneIDs <- getSig(cuff,level="genes",alpha=0.05)
#Use returned identifiers to create a CuffGeneSet object with all relevant info for given genes
diffGenes<-getGenes(cuff,diffGeneIDs)
#gene_short_name values (and corresponding XLOC_* values) can be retrieved from the CuffGeneSet by using:
names<-featureNames(diffGenes)
row.names(names)=names$tracking_id
diffGenesNames<-as.matrix(names)
diffGenesNames<-diffGenesNames[,-1]
# get the data for the significant genes
diffGenesData<-diffData(diffGenes)
row.names(diffGenesData)=diffGenesData$gene_id
diffGenesData<-diffGenesData[,-1]
# merge the two matrices by row names
diffGenesOutput<-merge(diffGenesNames,diffGenesData,by="row.names")
Does anyone have a solution to this problem? Regards

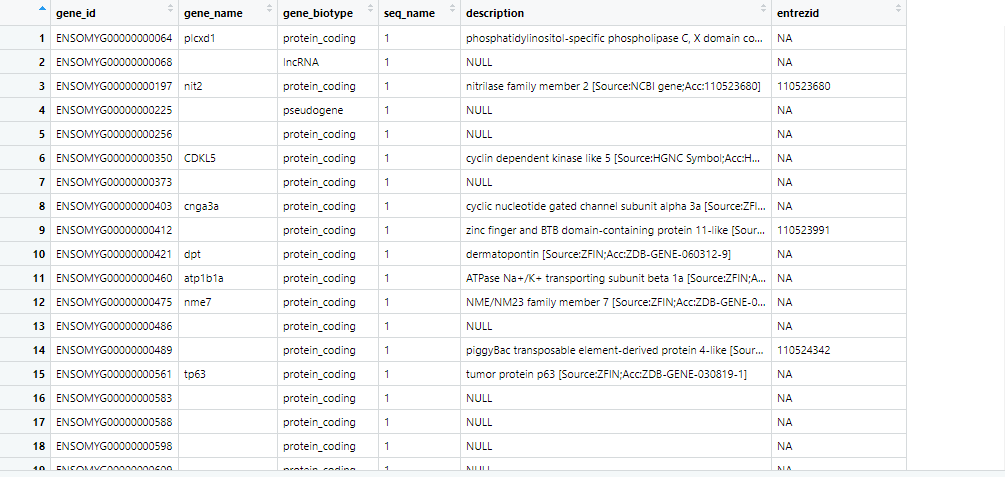

Dear Dr. Kevin Blighe, I would appreciate your immediate attention to this matter. Thank you very much for the prompt reply and your information. sincerely regards.
Try the solution by James, first. It will prove a lot easier.