Entering edit mode
I'm wondering is that possible to find if the sequence is enriched by specific motifs?
For example, I have generated Granges object and DNAStringset :
> gr_up <- GRanges(gr_up)
GRanges object with 376 ranges and 3 metadata columns:
seqnames ranges strand | ENSG pval dPSI
<Rle> <IRanges> <Rle> | <character> <numeric> <numeric>
1 chr1 167012370-167012510 + | ENSG00000143194 0.0000000 1.000000
2 chr1 186304109-186304256 + | ENSG00000116690 0.0874126 0.666667
3 chr1 154176116-154176178 - | ENSG00000143549 0.0000000 0.488313
4 chr1 203347293-203348276 - | ENSG00000122176 0.0969031 0.482400
5 chr1 54017377-54017426 - | ENSG00000203985 0.0289710 0.433965
... ... ... ... . ... ... ...
634 chrX 38804311-38804412 + | ENSG00000165175 0.0974026 0.134225
635 chrX 68511850-68511976 + | ENSG00000181704 0.0309690 0.128942
636 chrX 48575562-48575629 + | ENSG00000102317 0.0299700 0.119535
637 chrX 47583409-47583535 + | ENSG00000102265 0.0109890 0.115762
638 chrX 136548590-136549007 + | ENSG00000102243 0.0559441 0.100380
-------
seqinfo: 23 sequences from an unspecified genome; no seqlengths
> seqs_up <- getSeq(BSgenome.Hsapiens.UCSC.hg38, gr_up)
DNAStringSet object of length 376:
width seq names
[1] 141 TCCCCTAGTCTCCTGATGCTTCTTGTCATAATTCTTCTTGGACTAATTCACTG...TAATCAACAAATATTTATTGAGCACCTCCTCTGTGCCAGAAGATGATCCAAA 1
[2] 148 GCATAATCCCACATCACCACCATCTTCAAAGAAAGCACCTCCACCTTCAGGAG...ACCAAACAAGAAGAAGACTAAGAAAGTTATAGAATCAGAGGAAATAACAGAA 2
[3] 63 AGAGTGAGAGTAGTCGTCGAAAAAGTCGAAGAAGGTCGAAAACGTCCCGTCACCGGTCCGCGA 3
[4] 984 TAACTAGGATAACGGAACCTCCATCTCCAAGAGGTCCAACCACAACTGACCCC...TCTCCCTCTTCTCGGGACGGTCGTCGTCCTCCCTCCAGGTGACGTAAAACAG 4
[5] 50 AAGGGGAGGTCCCACCCGAAACCTAAGTCGTCGTCACATCGGTAAGAGAG 5
... ... ...
[372] 102 CACAGTCCAGCCACTGACCGCAGCAGCGCCCTTGCGTAGCAGCCGCTTGCAGCGAGAACACTGAATTGCCAACGAGCAGGAGAGTCTCAAGGCGCAAGAGGA 634
[373] 127 TTGCAGGCCTTTCAGATATATCCATCTCACAAGACATCCCCGTAGAAGGAGAA...GCATCCGGGAGTTTGACAGCTCCACATTAAATGAATCTGTTCGCAATACCAT 635
[374] 68 GGTCGTTGTCAAGGACCGGGAGACTCAGCGGTCCAGGGGTTTTGGTTTCATCACCTTCACCAACCCAG 636
[375] 127 ACCCACCATGGCCCCCTTTGAGCCCCTGGCTTCTGGCATCCTGTTGTTGCTGT...CTGCACCTGTGTCCCACCCCACCCACAGACGGCCTTCTGCAATTCCGACCTC 637
[376] 418 TGATAGCATGTCTCCAAATCAGTGGCGTTACTCGTCTCCATGGACAAAGCCAC...GATAGCTGGAAGCACAGGGTTGCTCTTCAACCTGCCTCCCGGCTCAGTTCAC 638
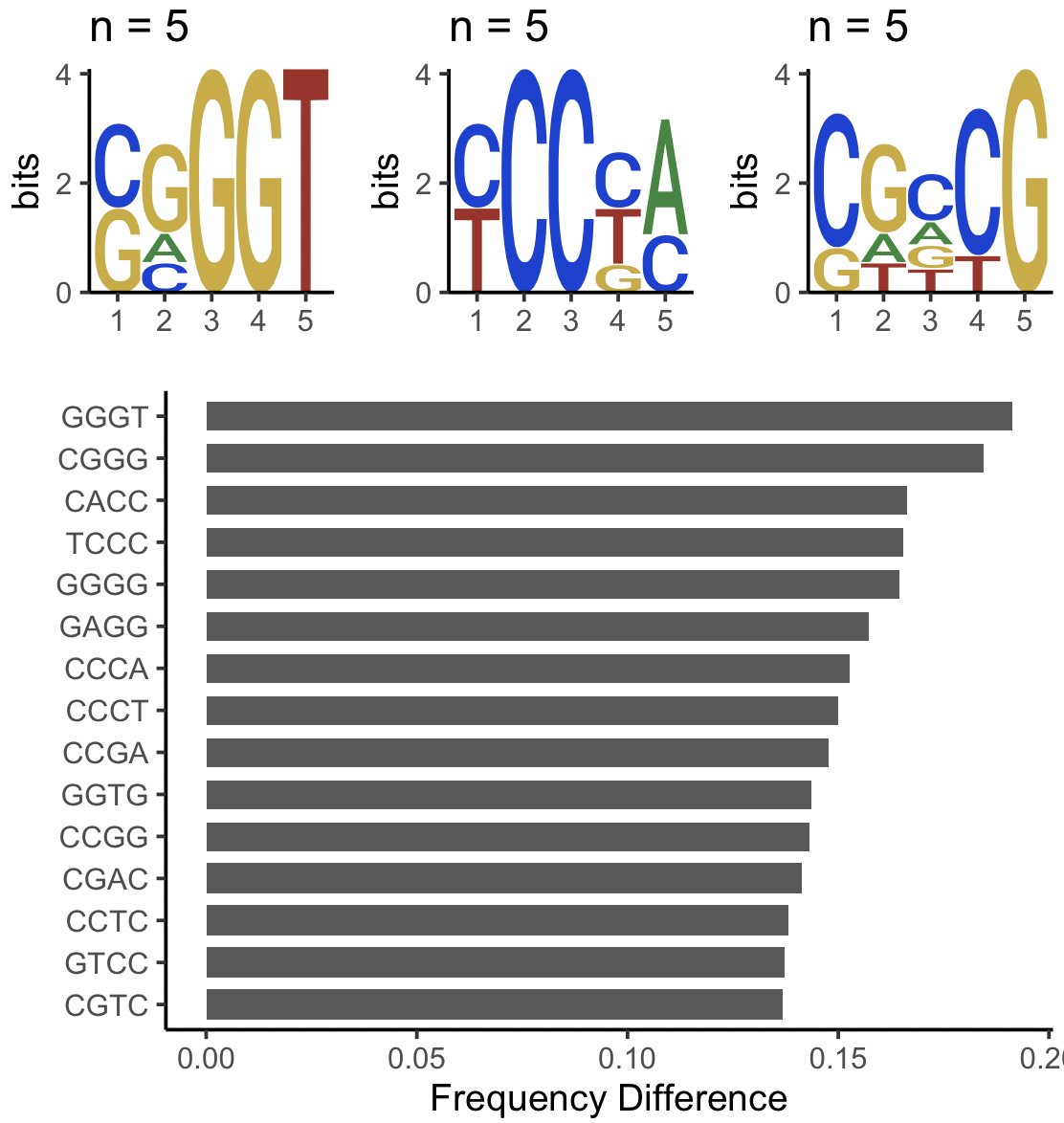
And then I have conducted motif enrichment analysis and found that these sequence set have some "C" enrich.
I'm wondering is that possible to find which specific sequence(in Granges or DNAStringset ) has "C" enrich?