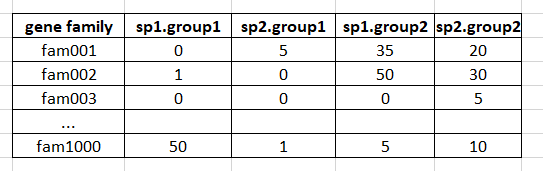
Hi,
Since edgeR appears to be designed to deal with count values, I was wondering if it could be applied to roughly detect differences in gene counts in gene families of two groups, for example (numbers below represent counts):
I'm assuming that each species in such matrix could be considered a replicate within each group, that gene families would be the equivalent to loci, and counts are absolute.
Any suggestion/advice are will be welcome.
Eliza