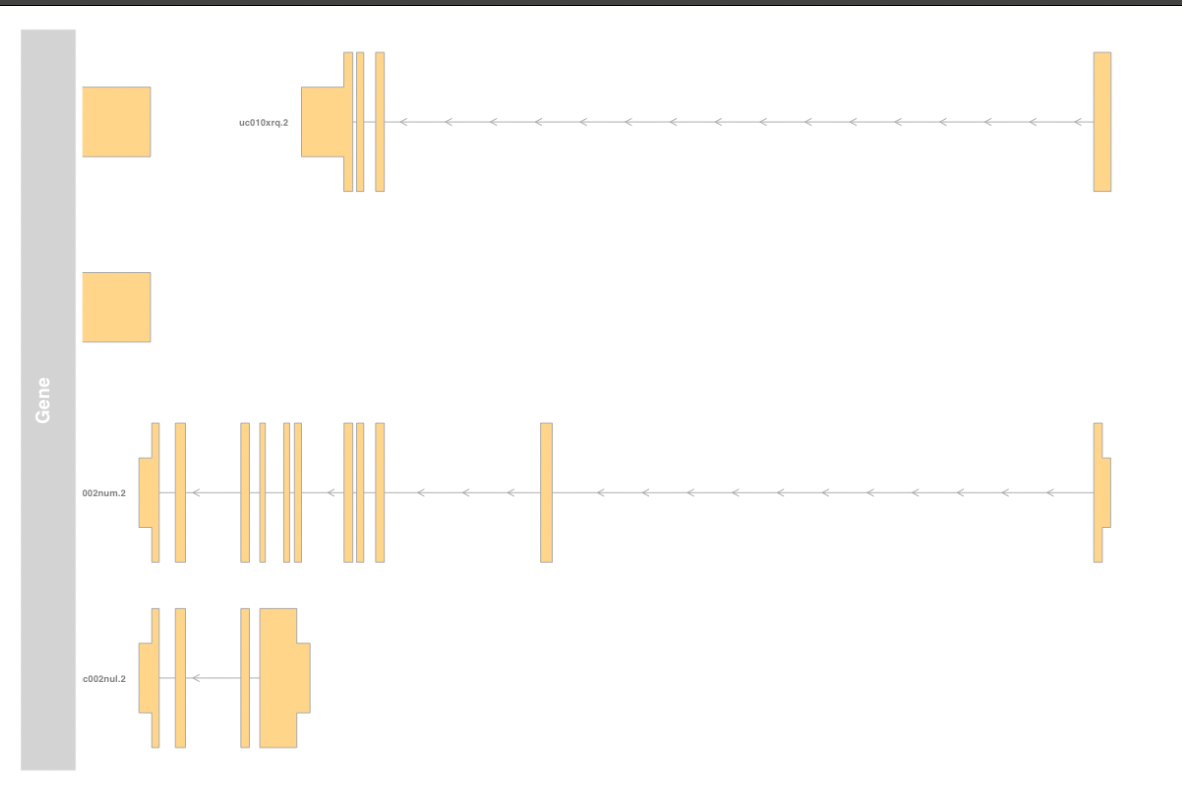
Entering edit mode
Hello. I am trying to create a GeneRegionTrack using Gviz. I would like to display the gene symbol rather than the Ensembl transcript ID in the gene track. Please see below:
gene <- GeneRegionTrack(TxDb.Hsapiens.UCSC.hg19.knownGene,
name ='Gene',
showId = TRUE,
transcriptAnnotation = "symbol",
geneSymbol = TRUE)
plotTracks(gene,
from = start,
to = end,
chromosome = 'chr19')
The main issue here is that the TxDb object does not contain information about the gene symbol.
columns(TxDb.Hsapiens.UCSC.hg19.knownGene)
How to add the info of gene symbols in the TxDb object, then show the GeneRegionTrack with gene symbol instead of only the Ensembl transcript's ID?