Hi,
I am working with microRNA microarray data and I am facing very strong batch effetcs. In particular, I have 15 samples with a disease and 15 healthy controls. The first thing that I did was to explore the variance of miRNA expression by PCA/MDS plots and i immediately noted the presence of multiple confounding factors. Later, i looked into detail at the sources of batch effects with a PVCA, which reported that the main source of variability is due to the day on which the samples were analyzed, the array lot number and the RNA extraction date.
Therefore, i included these factors (together or singularly) in the design matrix in limma, but, unfortunately, no differentially expressed genes were found, even though i am sure that biologically explained differences are present. I've also tried to include arrayweights (as i am working with clinical samples) and i've also removed lowly expressed genes.
Furthermore, i've also tried to correct expression data directly with ComBat. In this case, i am able to spot few differences when correcting for lot number, whereas i find big differences when correcting for the date. However, i heard that ComBat on unbalanced designs might result in too many significant genes, and that, generally, including the confounding variable in the linear model is better than directly changing expression values. I have also tried with SVA but few things changed.
The lot number is distributed between samples as follows:
| Controls | Disease | |
|---|---|---|
| Lot 1 | 3 | 4 |
| Lot 2 | 9 | 11 |
| Lot 3 | 1 | 0 |
Instead, the date of the experiment is unbalanced:
| Controls | Disease | |
|---|---|---|
| Day1 | 0 | 1 |
| Day2 | 0 | 1 |
| Day3 | 0 | 1 |
| Day4 | 1 | 0 |
| Day5 | 2 | 0 |
| Day6 | 2 | 3 |
| Day7 | 1 | 1 |
| Day8 | 1 | 2 |
| Day9 | 1 | 1 |
| Day10 | 0 | 1 |
| Day11 | 1 | 3 |
| Day12 | 1 | 0 |
| Day13 | 0 | 1 |
| Day14 | 1 | 0 |
| Day15 | 2 | 0 |
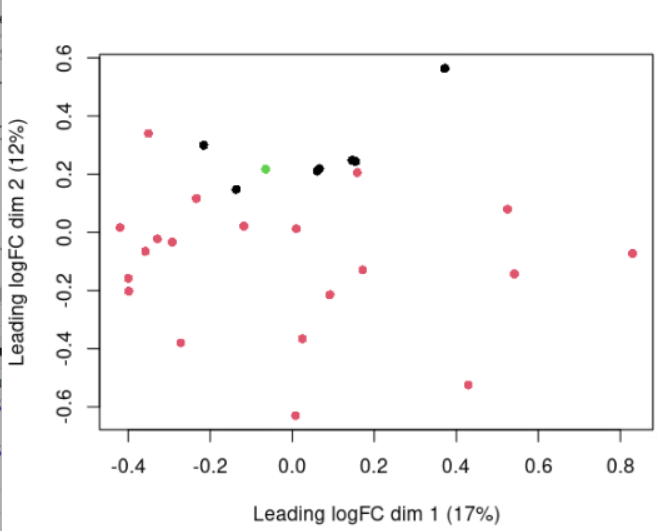
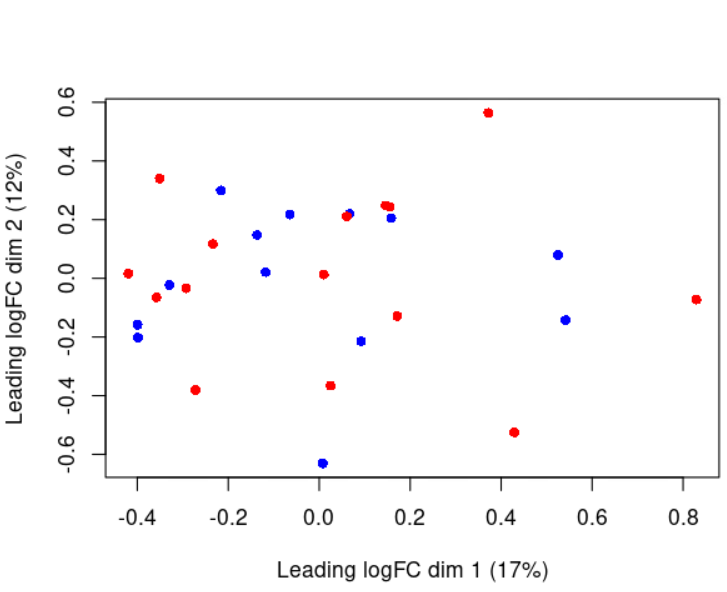
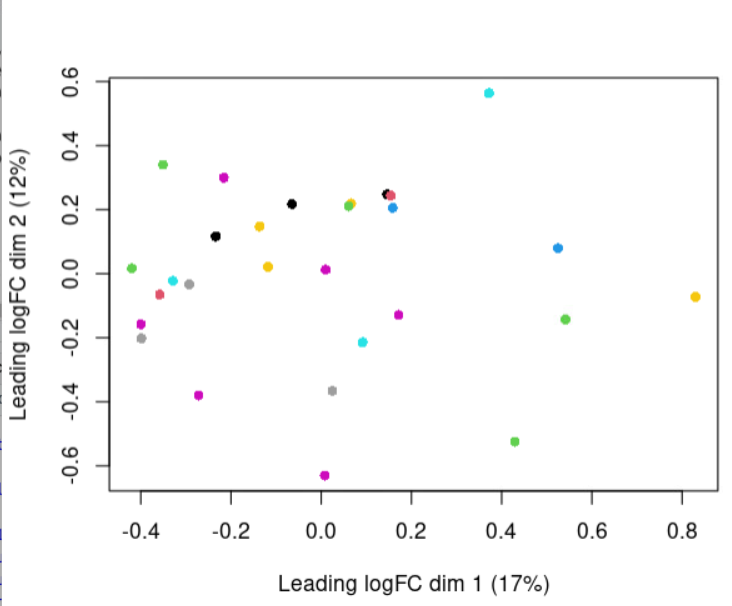
Moreover, i report the MDS plot colored by disease, experiment date and lot number, respectively:
How should i proceed? Is there something that i can do with this number of samples to correct these batch effects? Thanks in advance!