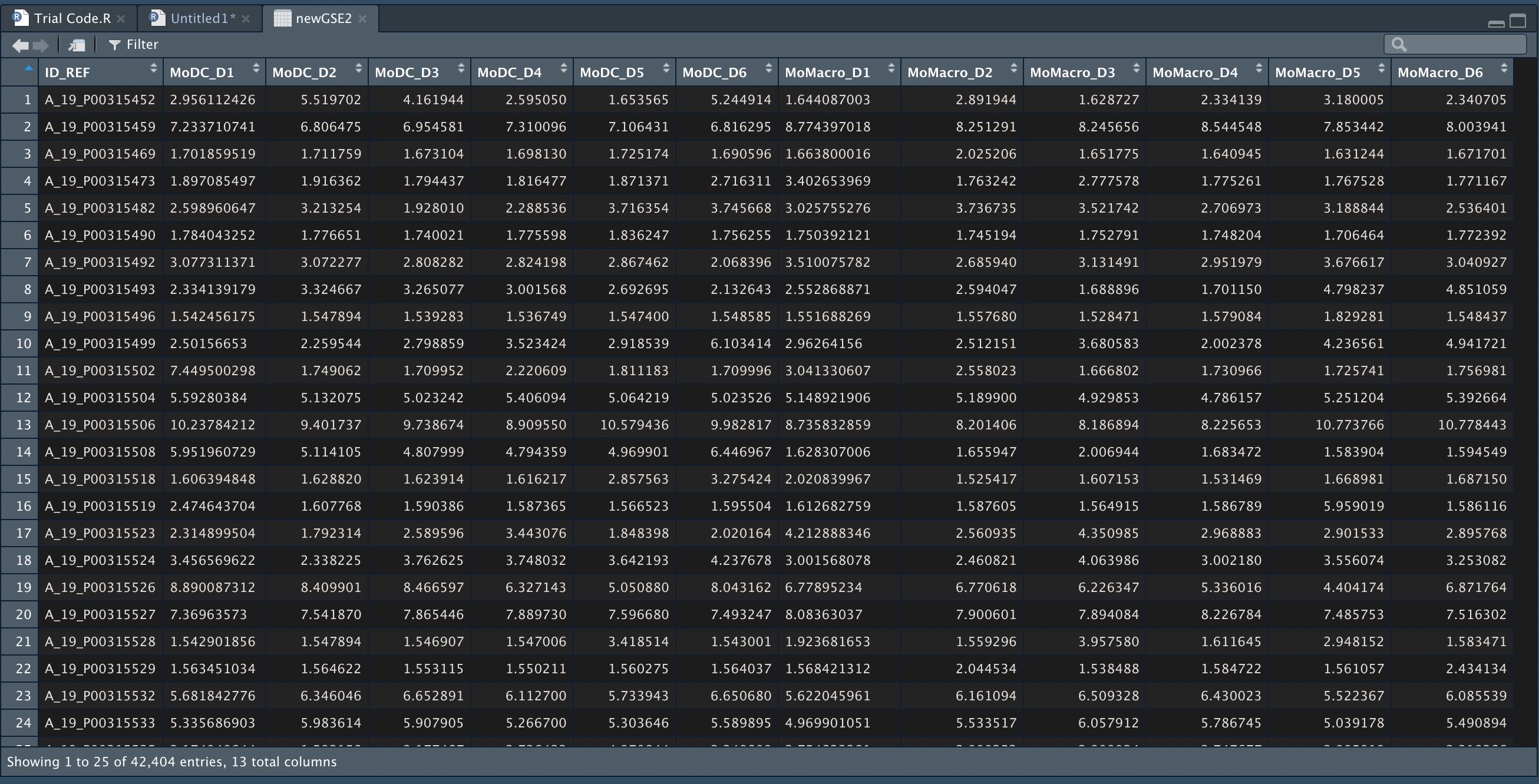
Hi, I am quite new to R and I have been trying to use a dataset from the GEO database (GEO104438) that is interesting to my PhD work. The file that I have been able to download and use in R, is a table with ID references and then the normalised signal intensity values for all of the donors (and cell types). I have put a screenshot of the table for reference.
I am hoping to try look at whether there are any genes that are differentially expressed depending on what cell type they are (as they are both derived from a common precursor). So for each donor I have 2 different cell types. Due to this, I was initially trying to tell R that my data is paired but I am stuck.
I have been reading online and trying to use the limma guide but I am completely confused as all of the examples I have seen online are not in a similar format to this dataset.
I would appreciate any help!


Thank you so much!
Also, I didn't realise it was easier to use GEOquery rather than actually reading the data file in - I will definitely remember that in future.
You can also use
GEOqueryto get the raw data if available. Unfortunately, what the submitter uploaded as 'raw' data in this instance is not actually the raw data, so it's a moot point.So I have been working on this very slowly (its a side project), and working through understanding the code.
I am now at the line where we are making contrasts. When I run this code it is coming up with an error message saying "Error in eval(ej, envir = levelsenv) : object 'ATDC' not found".
I have been doing some reading and I think it is because my column name is Cell, and then ATDC/MoDC/MoMacro are under this. But I am unsure how to fix this.
Using the limma users guide, I tried running the following code but that didnt seem to help either:
Your goal when asking questions should be to provide all the information possible about what you were doing when you got the error. Telling us what you did that didn't work is not useful. Ideally you would run your code and then post enough of the code that others could replicate, like what I did above, as well as the error message, and probably the output from running
traceback()right after the error.Hi James, as I mentioned that I had used the code you previously provided to help (found above) I didn't think to re-print the code. I have now provided it below:
As I receive the error message at the point of code where we try do the contrasts, I am unable to go any further. As I mentioned, I tried reading and trying some other lines of code but these are not working (so I will not repeat them so as to not cause any further confusion).
As per your suggestion, I ran the traceback code too, which I have put below:
As I am still getting the hang of R, I would appreciate any help!
Ah, my fault. Look at the colnames of your design. You will need to do one extra step
And now the column names of your design should be OK. But check!