Entering edit mode
Following the instructions to install clusterProfiler I did
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("clusterProfiler")
sessionInfo( )
R version 4.0.5 (2021-03-31)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=Spanish_Spain.1252 LC_CTYPE=Spanish_Spain.1252
[3] LC_MONETARY=Spanish_Spain.1252 LC_NUMERIC=C
[5] LC_TIME=Spanish_Spain.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods
[7] base
loaded via a namespace (and not attached):
[1] tidyselect_1.1.2 purrr_0.3.4 haven_2.4.1
[4] lattice_0.20-41 carData_3.0-4 colorspace_2.0-1
[7] vctrs_0.3.8 generics_0.1.3 htmltools_0.5.1.1
[10] utf8_1.2.1 rlang_1.0.6 pillar_1.7.0
[13] foreign_0.8-81 glue_1.4.2 DBI_1.1.3
[16] readxl_1.3.1 lifecycle_1.0.1 munsell_0.5.0
[19] gtable_0.3.1 cellranger_1.1.0 zip_2.1.1
[22] htmlwidgets_1.5.4 leaps_3.1 rio_0.5.29
[25] forcats_0.5.1 curl_4.3.1 fansi_0.4.2
[28] broom_0.7.6 Rcpp_1.0.6 DT_0.22
[31] flashClust_1.01-2 backports_1.2.1 scales_1.2.0
[34] BiocManager_1.30.19 writexl_1.4.0 scatterplot3d_0.3-42
[37] abind_1.4-5 digest_0.6.27 ggplot2_3.3.5
[40] hms_1.1.1 stringi_1.5.3 openxlsx_4.2.3
[43] rstatix_0.7.0 dplyr_1.0.6 ggrepel_0.9.1
[46] grid_4.0.5 cli_3.4.1 tools_4.0.5
[49] magrittr_2.0.1 tibble_3.1.1 cluster_2.1.1
[52] crayon_1.5.2 car_3.0-10 tidyr_1.1.3
[55] FactoMineR_2.4 pkgconfig_2.0.3 ellipsis_0.3.2
[58] MASS_7.3-53.1 data.table_1.14.0 assertthat_0.2.1
[61] rstudioapi_0.14 R6_2.5.1 compiler_4.0.5
I don't really know which is the problem why I can't install the package It takes hours to install with infinite lines of code (I have made screenshot of several) getting errors of lazy loading along to indecipherable code.enter image description here
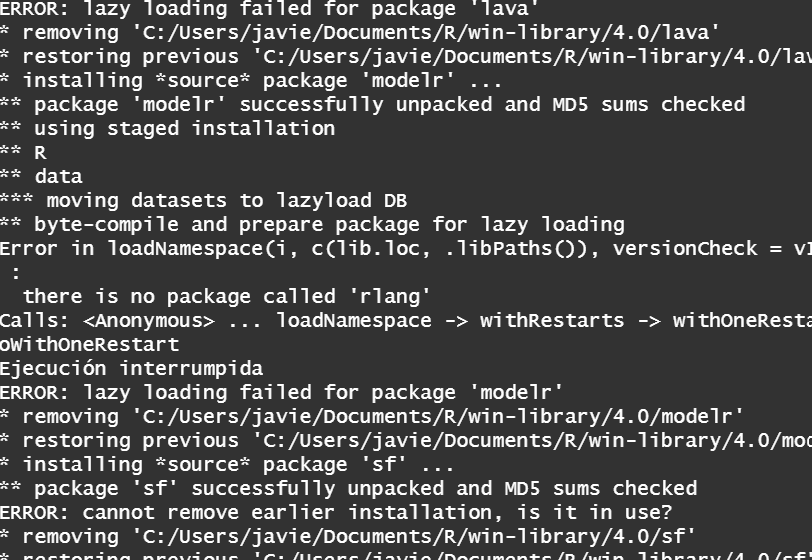

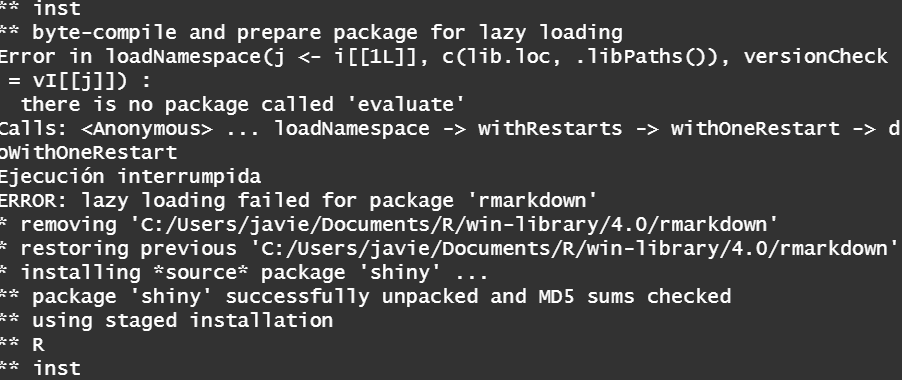

install.packages("rlang")and reload your R session.... and is there a reason you are not using the latest version of R? That is meanwhile version R-4.2.1!
If there is no such reason, you should install the latest version...
I went to check updates and I get: "You're using the newest version of RStudio"
I am not sure if this means that I have the last version, but it's true that getting the R version is 4.0.5 You recommend download the new version?
Please realize that RStudio is something else than R! So in addition to RStudio, you should also have R installed. And updating RStudio does not update R,
Preferably you use the latest versions of both, but you can have multiple instances (versions) of R installed on your computer. Within Rstudio you can select which version of R to use.
But yes, also go for the latest version of R (you apparently already have the latest version of RStudio installed). Be sure, though, to 'instruct' RStudio to use R-4.2.1.