Entering edit mode
I'm looking into the differentially expressed genes in heat-stressed animals versus normal ones.
When I only done the analysis on the 6 samples (3 heat stressed and 3 non-stressed) I got a few significant genes that are significantly differentially expressed on stress.
When I did the DEseq analysis using data matrix of all samples that include RNA-seq of 40 samples 3 of them are heat stressed the other 3 are non-stressed and the rest are of different other conditions like ageing and cold stress.
dds <- DESeqDataSetFromMatrix(countData = data,
colData = samples,
design = ~0+condition)
and in my DEseq contrast
ageing <- results(dds_lrt,
contrast = c("condition", "5.months.old", "young"))
stressed <- results(dds_lrt,
contrast = c("condition", "heat-stressed", "control"))
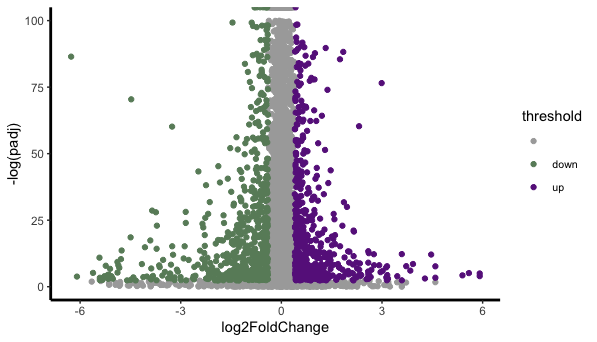
by applying that I got hundreds of significant genes.
Is this a right way of doing it and the volcano blot is weird