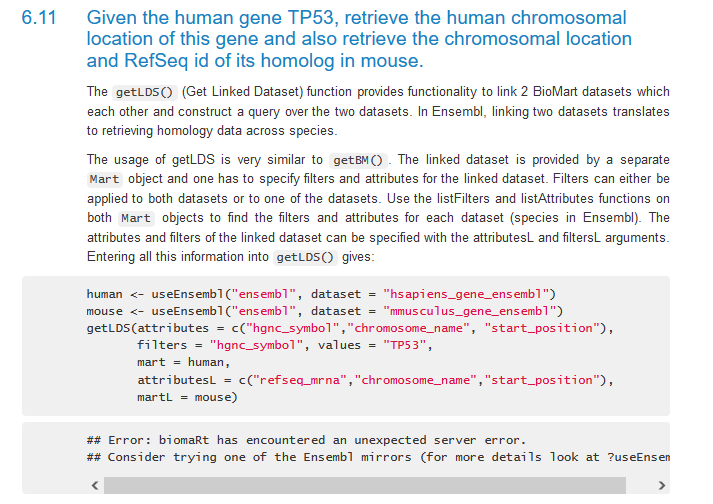
Entering edit mode
Hi,
I want to retrieve human and rat homologs from the Ensembl 107 archive and I get the following error:
mart_rat <- useMart(host='jul2022.archive.ensembl.org',biomart="ENSEMBL_MART_ENSEMBL",dataset="rnorvegicus_gene_ensembl")
mart_human <- useMart(host='jul2022.archive.ensembl.org',biomart="ENSEMBL_MART_ENSEMBL",dataset="hsapiens_gene_ensembl")
# get homology information
bmh <- getLDS(attributes = c("ensembl_gene_id", "external_gene_name"),
mart = mart_human,
attributesL = c("ensembl_gene_id", "external_gene_name"),
martL = mart_rat
)
## Error: biomaRt has encountered an unexpected server error.
## Consider trying one of the Ensembl mirrors (for more details look at ?useEnsembl)
It turns out that the same error is documented in the package vignette from 11.22...
This makes me think that the problem does not come from my code... I should also say that I tried it with different annotation versions and I encounter the same problem from Ensembl 106 to the current version.
Any solution from your side?
Many thanks!