When I tried to create a list with EdgeR, I encountered a error:"Error in DGEList(counts = cnt, group = group) : non-numeric values found in counts" . I could not solve this error and I hope you can help me:
SessionInfo():
Data base information:
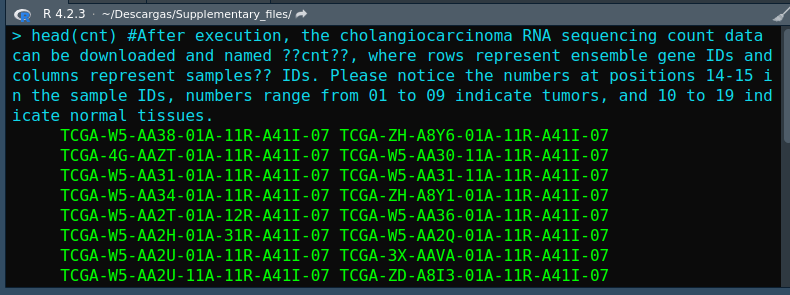
I hope you can help me to find out what the error is and how to solve it.

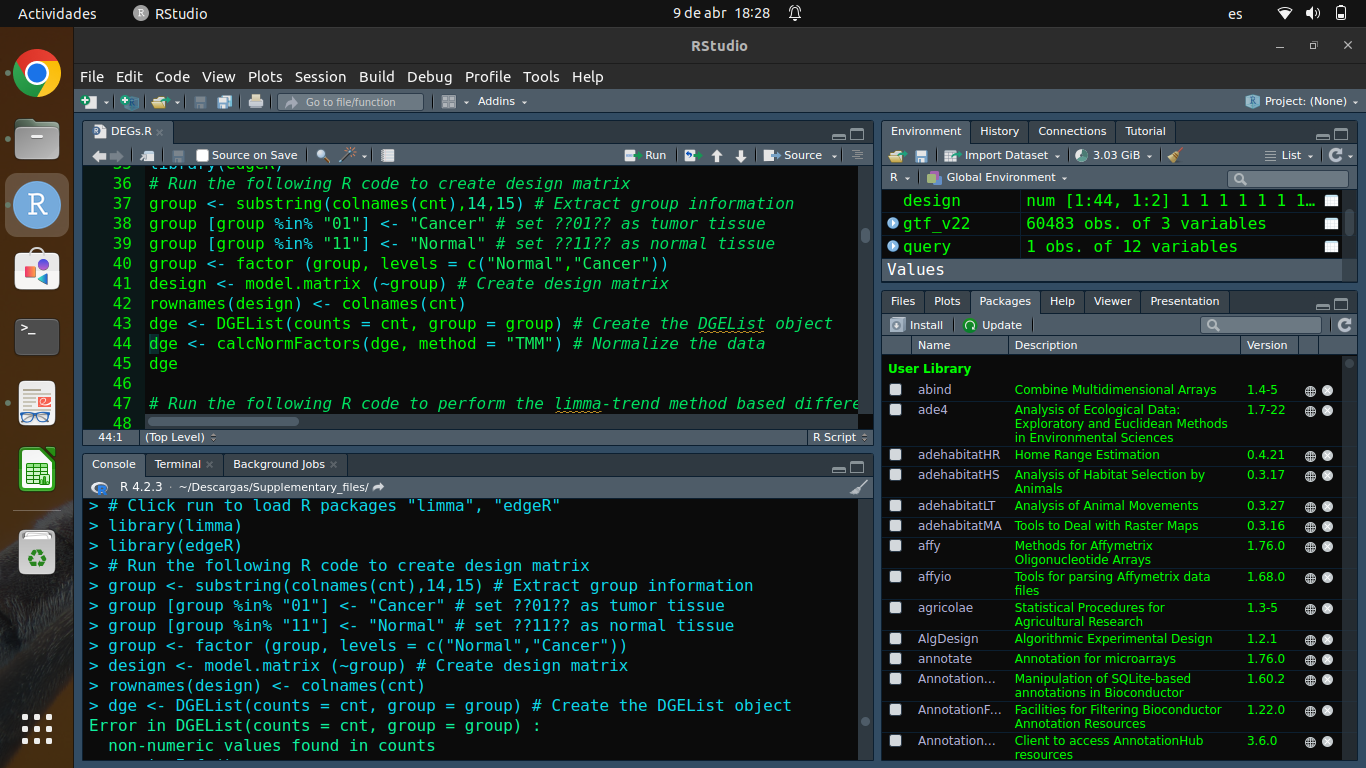

Please show code and output as text rather than as screen shots. The error message concerns
cnt, but you haven't shown the code by whichcntwas created, so we can't tell you how to solve the problem.cnt:
All pipeline: