Entering edit mode
Hi,
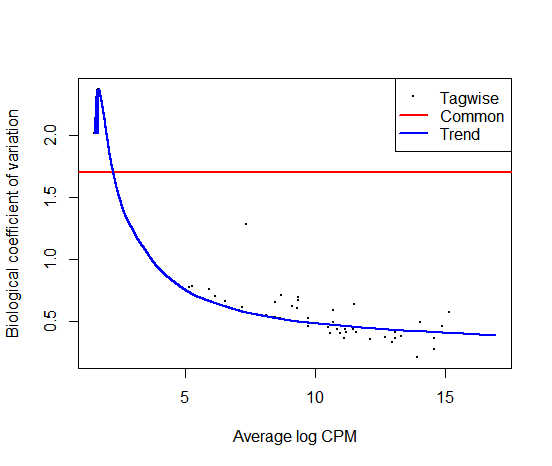
Based on the observations from the plotBCV, the common biological coefficient of variation (BCV) for my count data is around 1.7. However, there are not many genes with a similar BCV value. I am wondering whether I should manually adjust the BCV value to 0.5 or 0.6 for the exactTest function in edgeR.
Is it advisable to manually adjust the BCV value in this situation? If so, how should I choose the new value?
Also, to calculate the BCV from the common.dispersion value in edgeR, the formula is simply bcv = sqrt(edgeR_obj$common.dispersion)?
Any advice would be appreciated.