Hello guys,
I am doing some RNA-seq analysis using DESeq2 and following the vignette from here, where they use time points 0,15,30,60 ... 180 min. and two groups (strains)
http://www.bioconductor.org/help/workflows/rnaseqGene/
In the time series chapter it says:
"Genes with small p values from this test are those which, at one or more time points after time 0 showed a strain-specific effect."
As an example the following is shown:
## log2 fold change (MLE): strainmut.minute180
## LRT p-value: '~ strain + minute + strain:minute' vs '~ strain + minute'
## DataFrame with 4 rows and 7 columns
## baseMean log2FoldChange lfcSE stat pvalue padj symbol
## <numeric> <numeric> <numeric> <numeric> <numeric> <numeric> <character>
## SPBC2F12.09c 174.6712 -2.65763737 0.7498270 99.23199 7.671942e-20 5.184698e-16 atf21Question 1)
As I am understanding it- this gene has a strain specific expression profile over time. This effect occures over the other time points (from 0 to 180).
If I had picked the strainmut.minute60 then we would have seen a list of DE genes from 0 to 60 minutes, right?
Just for clarification- here are the options of which one can choose:
resultsNames(ddsTC)
## [1] "Intercept" "strain_mut_vs_wt" "minute_15_vs_0" "minute_30_vs_0"
## [5] "minute_60_vs_0" "minute_120_vs_0" "minute_180_vs_0" "strainmut.minute15"
## [9] "strainmut.minute30" "strainmut.minute60" "strainmut.minute120" "strainmut.minute180"Question 2)
Imagine a gene that behaves in the same way in all timepoints (between the two groups) but time point 0 to 15.
Will this be detected by deseq?
Cheers,
john

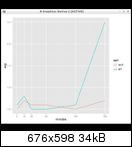

See the DESeq2 vignette for details on how to tell R which level is the reference level for a factor variable.
In this case the reference level was set during object construction which can be followed at the fission package vignette.
Is there a way to take all time points into account? In order to find a gene that is DE in all time points between two groups?
If you want to find genes which have a significant mut vs wt fold change for each and every time point, I don't think there is a simple way to form this results table. You would have to test the fold change for mut vs WT at minute 0:
...and then test the fold change for mut vs wt at every other time point:
And you would then look for genes which have a small adjusted p-value in all of the results tables.
To clarify: the first comparison takes into account the difference at t0, and this is recommended. The second doesn't take into account the difference at t0, and this is not usually recommended.
So if we do
results(dds, contrast=list(c("strain_mut_vs_wt","strainmut.minute15")))and then
results(dds, contrast=list(c("strain_mut_vs_wt","strainmut.minute30")))and on and on and then just select the genes that have an adjusted pvalue less than some threshold for all these tests, would this give the correct significance? I mean all of these tests share strainmut.minute0 as the control so they are correlated. How can we model this correlation? Instead, is there a way to test for a composite hypothesis such as \beta_1 >0 AND \beta_2 >0 where \beta_1 and \beta_2 are two different fold change effects with the same baseline?
This isn't a concern in linear models, think about the coefficient table that lm() returns for example when there are 3 or more factor levels. The assumption is that the data is not correlated conditioned on the design matrix X.
No, we don't have a way to test such composite alternative hypotheses.
I am interested in genes that are DE at each time point (i.e. ALL time points) with respect to 0. I ran some simulations for the lm model at http://rpubs.com/ysdel/corpvals and it seems that the tests are correlated. I don't know how to think about this correctly to find the significance levels.
I don't really follow your question, because I don't see where we make the assumption that test statistics from different contrasts would be independent.
Nevertheless, we don't have in DESeq2 a way for you to generate a p-value where the alternate hypothesis is that all time points after t0 have consistent sign of LFC compared to t0.
I'm sorry, I wasn't clear in what the problem is. Suppose I have results res1, res2, res3 from correlated tests. And I am interested in genes that satisfy the alternative hypothesis for all of these tests. So I will have to do
hits = (res1$padj<0.05) & (res2$padj<0.05) & (res3$padj<0.05)[or, forgetting about multiple testing adjustments,
hits = (res1$p.value<0.05) & (res2$p.value<0.05) & (res3$p.value<0.05)]What is the statistical significance (in terms of FDR or Type I errors) of these hits? If I had a simple hypothesis, I know for (res1$padj<0.05) I will make a type I error 5% of the time.
I very much appreciate you taking the time to answer these questions.
You're correct that that statement doesn't have the same control as the tests we provide. We don't have functionality for this kind of combination in DESeq2. You could use pvalue combination methods.
Hi Michael,
I am analyzing RNA-Seq data from a time-course of a mutant strain and would like to ask you something related to this reply.
Following what is described in the time-course vignette:
- In order to calculate the fold change for mut vs wt at an individual time-point shouldn't we use the Wald test like in:
What is the difference between using the one above and the following contrast, which as you describe, "(...) test the fold change for mut vs wt at every other time point" (time-point 15 in this particular case):
results(dds, contrast=list(c("strain_mut_vs_wt","strainmut.minute15")))Many thanks!
Miguel
"strainmut.minute15" is the difference between Mut vs WT at minute 15, controlling for baseline. If you add "strain_mut_vs_wt" to this, you get the LFC for Mutant vs WT at minute 15, not controlling for baseline. So the second one is the observed difference at minute 15 between the two groups (because you added in the change that was present at time=0).
Hi Michael, Just went through this post and I have a question. I have an experiment with 3 conditions (let's called them A/B/C) at 3 different time-points (0h/12h/24h) Following the tutorial I set
dds<-DESeqDataSetFromMatrix(countData = matrix, colData = colData, design = ~ condition + time + condition:time) dds <- DESeq(dds, test="LRT", reduced=~condition+time)
Thus, running
will give me the genes specifically DE at 12h, regarding if they are or are not DE at 0h I guess the same apply for 24h, that is, using
will give me genes specifically DE at 24h regarding if they are or are not DE at 0h AND 12h, right?
Now, to have genes that are DE ONLY at 0h, should I just use
"condition_A_vs_B"? Moreover, what if I need to find genes that are DE in one condition (say condition A) between different time points (e.g. 12h vs 0h) ?Thank you
Dave
This gives the condition effect at time 12 and 24 hrs, respectively.
If you want DE at a time point, and not DE at other time points, you can intersect gene lists.
In order to define "not DE", I recommend using
lfcThreshold > 0andaltHypothesis="lessAbs".Thank you Michael. Regarding the the first question I mispelled it, I meant "to have genes that are DE at time 0h" without "ONLY" Should then
"condition_A_vs_B"works?Regarding comparison of 2 condition at a specific time-point, If I would extract, from the whole expression matrix, a subset containing only samples at that specific time-point and run "standard" DESeq using
contrast=c("condition", "A", "B"), would I have the same results?Thank you again
Dave
Yes.
Similar but not same, see vignette FAQ.
Hi Michael,
I run the comparison of condition A vs B at all the 3 time-points:
Now, looking at the top genes, I get exactly the same values in all the 3 time-points, except for the log2FC and lfcSE which is odd...
For example:
Any clue why I get the same values for baseMean/stat/pval/padj ?
Thank you
Dave
If you have specified an LRT, you'd need to use
test="Wald"here to perform Wald tests of your contrasts.Hi Michael, sorry to bother you but going back to this post I realized I still need an answer to my question:
"What if I need to find genes that are DE in one condition (say condition A) between different time points (e.g. 12h vs 0h) ?"
I would really appreciate if you could help me Thank you
Dave
I think this is covered in the workflow:
https://bioconductor.org/packages/release/workflows/vignettes/rnaseqGene/inst/doc/rnaseqGene.html#time-course-experiments
Thank you Michael and sorry for my late reply but I got stuck on another project and forgot to reply. The workflow in the "Time course" experiment" it doesn't seem to answer my question (or at least I don't get it...). I read that the test showed in the text reports those genes that at some points after 0 show a strain specific effect. But how can I get genes that, in just one specific condition, change over time? Could I just use the read counts matrix of that specific condition in the 3 time points, use design = ~ time and then test between different time points? Thank you again
Dave
Start at the sentence: "Wald tests for the log2 fold changes at individual time points can be investigated using the test argument to results..."
For the reference condition you can just pull out results for e.g.
minute_15_vs_0withname(the vignette section on Interactions explains how this happens with a diagram).For non-reference condition, you just add the main effect
minute_15_vs_0and the interaction term, e.g.Got it Thank you very much
Dave
Hi Michael,
Just a quick question on the answer you gave for "Is there a way to take all time points into account? In order to find a gene that is DE in all time points between two groups?"
I followed your tutorial on the fission data for time series experiments, and i thought that taking a summary of results
> summary(resTC)after running the steps below would give an indication of the total genes differentially expressed across all time-points under consideration:Kindly clarify this part for me, because these are the same differentially expressed genes you then went on to use for construction of the heatmap
topGenes <- head(order(resTC$pvalue),40)So which time-point are these representing??
Thanks,
Stan
The likelihood ratio test here is testing genes for condition-specific differences over time after time 0. Here's the relevant section from the workflow:
"The following chunk performs a likelihood ratio test, where we remove the strain-specific differences over time. Genes with small p values from this test are those which, at one or more time points after time 0 showed a strain-specific effect. Note therefore that this will not give small p values to genes which moved up or down over time in the same way in both strains."