Entering edit mode
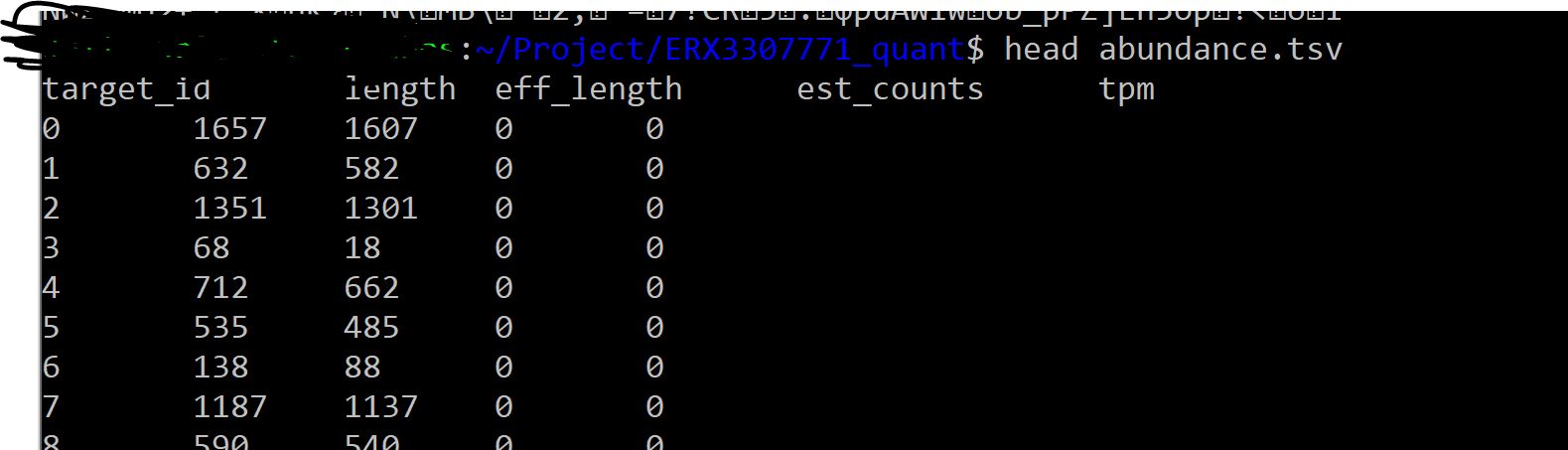
kallisto quant -b 10 -i Homo_sapiens_index -o ERX3307771_quant -t 1 --single -l 51 -s 1 -g Homo_sapiens.GRCh38.101.gtf ERX3307771.fastq
I think we need those target IDs to be not 0 1 2 3... in order to be able to apply tximport.
Does someone know what could be wrong here?
(I'm very new to bioinformatics)
Kindest regards Julie

