I am trying to run the following analysis in DESeq, and running into a few challenges, as described.
Subjects in two groups ('A', 'B') - with approximately 3 times more subjects in group 'A' vs. 'B', provide samples pre-treatment and at three time points post-treatment (t0,t1,t2,t3). Thus, our column data (with a nested index) looks something like:
We want to compare how the treatment differentially impacts subjects from group B vs. subjects from group A at any time post-treatment.
The first challenge is to create a DESeqDataSetFromMatrix because the design function
~ Group + Time + Group:Subject.nested + Group:Time gives an error that the 'model matrix is not full rank'. I plan to address this by using a custom model matrix for DESeq, but I need to create a DESeqDataSet object first - so I run DESEQDataSetFromMatrix call using 'ignoreRank=True'.
Then, following [1a,1b], I create a model.matrix (m1) with the same design as above, and then remove the zero columns.
Nevertheless, when I try to run DESeq(dds, full=mm1, betaPrior=FALSE), I still get the error that the 'model matrix is not full rank'.
Is there something incorrect in my logic?
Also, I believe I need to also use 'reduce' in DESeq to ~ Group + Time in order to obtain the correct final model (as outlined in [2]). Can you please help to confirm, and to integrate with the above?
Thank you.
[1a] With DESeq2 "Not full rank" Error with design ~ line + time + condition

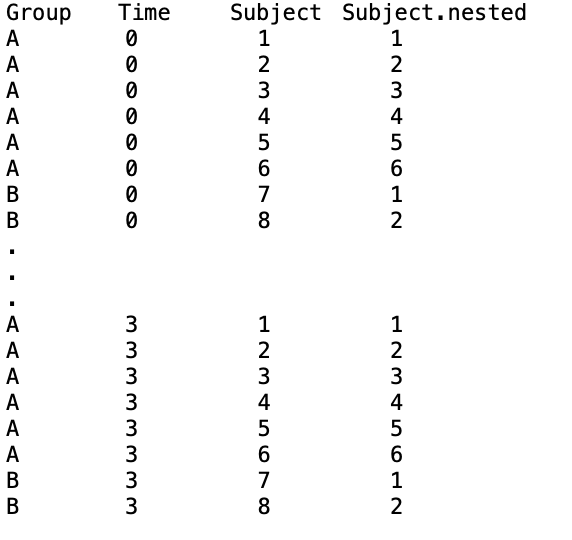

Michael, thank you very much for your response. Setting the design=~1 to create the original DESeqDataSet worked great to get around having to use ignorRank to initialize the object. Also, I found an error with my nested indexing, once I fixed it, I was able to run the full protocol to create a full-rank model matrix (m1) and run DESeq using 'full=m1'. I also found that I could run the reduced comparison by creating a model matrix for the reduced model (mr), and inputting that into the DESeq function (i.e. 'reduced = mr'). I will seek out further advice on appropriate construction of the design function.
Great. Sounds good.
I've also tried to clarify, in the vignette, the way to create the dataset for the case when the desired design leads to a less than full rank error:
https://github.com/mikelove/DESeq2/commit/f9aaacf110edda3f2c9d69b2fdbe94a633f41ecd