Hello,
I did a pseudotime analysis on a bulk RNA-seq data using phenopath. Here is the code I used to do this analysis:
x_mat <- model.matrix(~ Sex)
# sce is a SummarizedExperiment object containing expression as log2(TPM+1)
fit <- phenopath(sce, x_mat, elbo_tol = 1e-6, thin = 20)
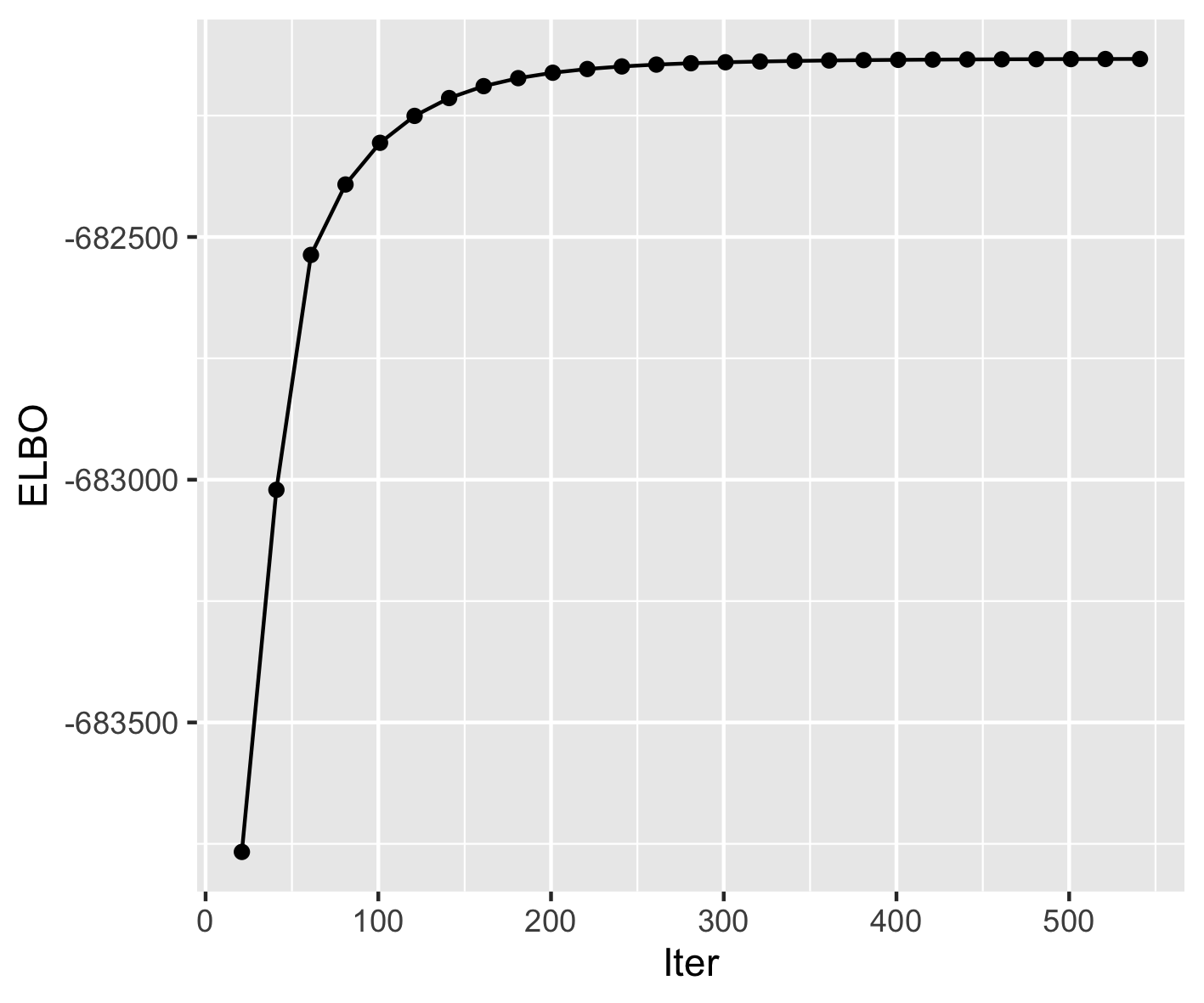
plot_elbo(fit)
I get the following elbo plot.
Now I would like to extract the information about the interaction between the latent space and my covariate (Sex).
I first tried to use the interactions function, and got an error:
> interaction_results <- interactions(fit)
Error: Assigned data `sig$feature <- chi$feature <- phenopath_fit$feature_names` must be compatible with existing data.
x Existing data has 2 rows.
x Assigned data has 2000 rows.
ℹ Only vectors of size 1 are recycled.
I then tried to extract the interaction information with the 3 following functions interaction_effects, interaction_sds and significant_interactions. Running these 3 functions, I was expecting to get vectors as stated in the vignette because I only have one covariate.
Note that if P=1 (ie there is only one covariate) each of these will return a vector, while if P>1 then a matrix is returned.
Instead, I get a matrix with 2 rows for the functions interaction_effects and interaction_sds, and 2 columns for the function significant_interactions. So I don't know how to interpret these results.
Could you help me get the interaction information from my analysis?
Thank you,
Clara Benoit-Pilven
> sessionInfo()
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.6
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats4 parallel stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 tidyr_1.1.3 dplyr_1.0.6 factoextra_1.0.6 ggplot2_3.3.3 ade4_1.7-16
[7] phenopath_1.10.0 SummarizedExperiment_1.16.1 DelayedArray_0.12.2 BiocParallel_1.20.1 matrixStats_0.55.0 edgeR_3.28.0
[13] limma_3.42.2 GenomicFeatures_1.38.2 AnnotationDbi_1.48.0 Biobase_2.46.0 GenomicRanges_1.38.0 GenomeInfoDb_1.22.0
[19] IRanges_2.20.2 S4Vectors_0.24.3 BiocGenerics_0.32.0
loaded via a namespace (and not attached):
[1] httr_1.4.2 bit64_0.9-7 assertthat_0.2.1 askpass_1.1 BiocFileCache_1.10.2 blob_1.2.1 GenomeInfoDbData_1.2.2
[8] Rsamtools_2.2.3 ggrepel_0.8.2 yaml_2.2.1 progress_1.2.2 pillar_1.6.1 RSQLite_2.2.0 lattice_0.20-40
[15] glue_1.4.2 digest_0.6.27 XVector_0.26.0 colorspace_2.0-1 htmltools_0.5.1.1 Matrix_1.2-18 XML_3.99-0.3
[22] pkgconfig_2.0.3 biomaRt_2.42.0 zlibbioc_1.32.0 purrr_0.3.4 scales_1.1.1 tibble_3.1.2 openssl_1.4.2
[29] farver_2.0.3 generics_0.1.0 ellipsis_0.3.2 withr_2.4.2 cachem_1.0.5 cli_2.5.0 magrittr_2.0.1
[36] crayon_1.4.1 memoise_2.0.0 evaluate_0.14 fansi_0.5.0 MASS_7.3-51.5 tools_3.6.2 prettyunits_1.1.1
[43] hms_0.5.3 lifecycle_1.0.0 stringr_1.4.0 munsell_0.5.0 locfit_1.5-9.1 Biostrings_2.54.0 compiler_3.6.2
[50] rlang_0.4.11 grid_3.6.2 RCurl_1.98-1.3 rstudioapi_0.13 rappdirs_0.3.1 labeling_0.4.2 bitops_1.0-6
[57] rmarkdown_2.8 gtable_0.3.0 DBI_1.1.1 curl_4.3.1 R6_2.5.0 GenomicAlignments_1.22.1 knitr_1.33
[64] rtracklayer_1.46.0 fastmap_1.0.1 bit_4.0.4 utf8_1.1.4 stringi_1.6.2 Rcpp_1.0.6 vctrs_0.3.8
[71] dbplyr_1.4.2 tidyselect_1.1.1 xfun_0.23