Hello, I am using the pattern search function of the Microbiome Analyst (microbiomeanalyst.ca) to compare abundance patterns in a metagenomics dataset (control vs treated).
A quick question: What is the difference between using 1-2 and 2-1 (1 being control and 2 treated) under PatternSearch>Predefined Profile? The results are slightly different, so could any of you give me more details on what are the implications? If I want to look for enriched taxa in my treated samples, which one should I choose?
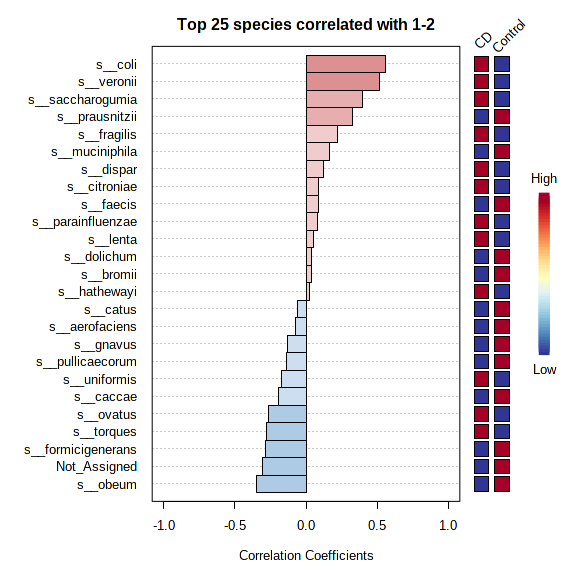
Another question. When running PatternSearch 1 vs 2, the graph shows the top 25 correlations. Are all red bars positive correlated with 2 and blue negative correlated with 1? Or are these correlations based on the side heatmap? Please see attached picture.
I know this may not be the best forum to ask, but I've contacted the developers via e-mail and their github forum, but could get an answer back! Thank you!