Entering edit mode
Hi,
I am trying to read a DBA object from a Samplesheet but DiffBind throws me an error despite having the file paths correctly set.
library(DiffBind)
setwd("C:/Users/alexa/Documents/HKU/diffbind_data/")
samples <- read.csv(file = "diffbind_design.csv")
samples <- dba(sampleSheet = samples)
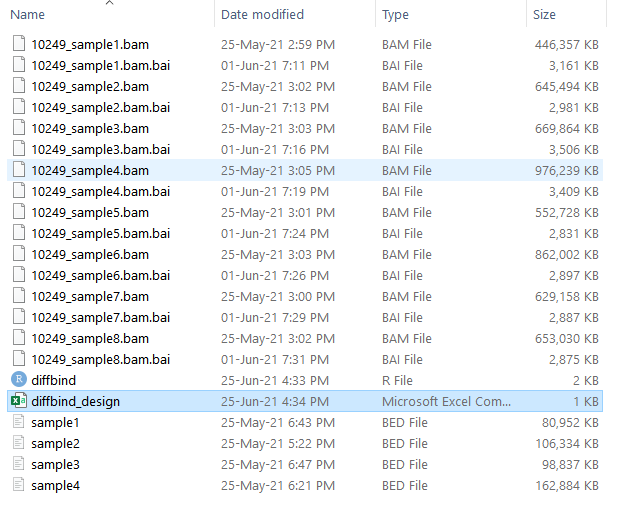
all files are under this directory C:/Users/alexa/Documents/HKU/diffbind_data/
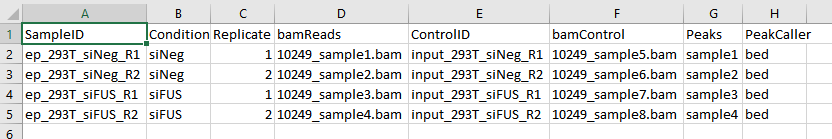
csv file looks like that:
sessionInfo()
R version 4.1.0 (2021-05-18)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] DiffBind_3.2.2 SummarizedExperiment_1.22.0
[3] Biobase_2.52.0 MatrixGenerics_1.4.0
[5] matrixStats_0.59.0 GenomicRanges_1.44.0
[7] GenomeInfoDb_1.28.0 IRanges_2.26.0
[9] S4Vectors_0.30.0 BiocGenerics_0.38.0
loaded via a namespace (and not attached):
[1] backports_1.2.1 GOstats_2.58.0
[3] BiocFileCache_2.0.0 plyr_1.8.6
[5] GSEABase_1.54.0 splines_4.1.0
[7] BiocParallel_1.26.0 ggplot2_3.3.4
[9] amap_0.8-18 digest_0.6.27
[11] invgamma_1.1 GO.db_3.13.0
[13] SQUAREM_2021.1 fansi_0.5.0
[15] magrittr_2.0.1 checkmate_2.0.0
[17] memoise_2.0.0 BSgenome_1.60.0
[19] base64url_1.4 limma_3.48.0
[21] Biostrings_2.60.1 annotate_1.70.0
[23] systemPipeR_1.26.2 bdsmatrix_1.3-4
[25] prettyunits_1.1.1 jpeg_0.1-8.1
[27] colorspace_2.0-1 blob_1.2.1
[29] rappdirs_0.3.3 apeglm_1.14.0
[31] ggrepel_0.9.1 dplyr_1.0.6
[33] crayon_1.4.1 RCurl_1.98-1.3
[35] jsonlite_1.7.2 graph_1.70.0
[37] genefilter_1.74.0 brew_1.0-6
[39] survival_3.2-11 VariantAnnotation_1.38.0
[41] glue_1.4.2 gtable_0.3.0
[43] zlibbioc_1.38.0 XVector_0.32.0
[45] DelayedArray_0.18.0 V8_3.4.2
[47] Rgraphviz_2.36.0 scales_1.1.1
[49] pheatmap_1.0.12 mvtnorm_1.1-2
[51] DBI_1.1.1 edgeR_3.34.0
[53] Rcpp_1.0.6 xtable_1.8-4
[55] progress_1.2.2 emdbook_1.3.12
[57] bit_4.0.4 rsvg_2.1.2
[59] AnnotationForge_1.34.0 truncnorm_1.0-8
[61] httr_1.4.2 gplots_3.1.1
[63] RColorBrewer_1.1-2 ellipsis_0.3.2
[65] pkgconfig_2.0.3 XML_3.99-0.6
[67] dbplyr_2.1.1 locfit_1.5-9.4
[69] utf8_1.2.1 tidyselect_1.1.1
[71] rlang_0.4.11 AnnotationDbi_1.54.1
[73] munsell_0.5.0 tools_4.1.0
[75] cachem_1.0.5 generics_0.1.0
[77] RSQLite_2.2.7 stringr_1.4.0
[79] fastmap_1.1.0 yaml_2.2.1
[81] bit64_4.0.5 caTools_1.18.2
[83] purrr_0.3.4 KEGGREST_1.32.0
[85] RBGL_1.68.0 xml2_1.3.2
[87] biomaRt_2.48.1 compiler_4.1.0
[89] rstudioapi_0.13 filelock_1.0.2
[91] curl_4.3.1 png_0.1-7
[93] tibble_3.1.2 stringi_1.6.1
[95] GenomicFeatures_1.44.0 lattice_0.20-44
[97] Matrix_1.3-3 vctrs_0.3.8
[99] pillar_1.6.1 lifecycle_1.0.0
[101] irlba_2.3.3 data.table_1.14.0
[103] bitops_1.0-7 rtracklayer_1.52.0
[105] R6_2.5.0 BiocIO_1.2.0
[107] latticeExtra_0.6-29 hwriter_1.3.2
[109] ShortRead_1.50.0 KernSmooth_2.23-20
[111] MASS_7.3-54 gtools_3.9.2
[113] assertthat_0.2.1 Category_2.58.0
[115] rjson_0.2.20 withr_2.4.2
[117] GenomicAlignments_1.28.0 batchtools_0.9.15
[119] Rsamtools_2.8.0 GenomeInfoDbData_1.2.6
[121] hms_1.1.0 grid_4.1.0
[123] DOT_0.1 coda_0.19-4
[125] GreyListChIP_1.24.0 ashr_2.2-47
[127] mixsqp_0.3-43 bbmle_1.0.23.1
[129] numDeriv_2016.8-1.1 restfulr_0.0.13