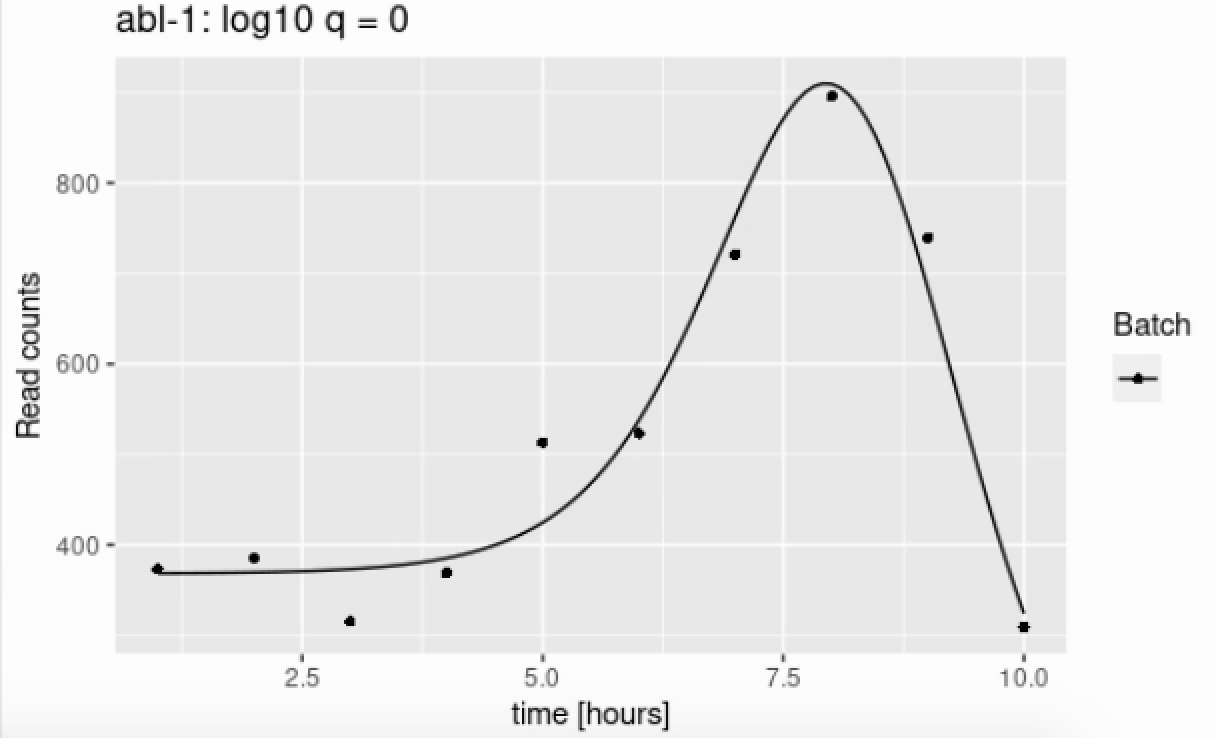
I am trying to use ImpulseDE2 to analyze time series RNA-seq data to identify monotonically and transiently expressed genes. From what I understand, the algorithm tries impulse, sigmoid, and constant fits in order to classify the genes. Below is an example of the gene-wise trajectory for abl-1:
When I set boolIdentifyTransients = TRUE, I get the following results:
impulseTOsigmoid_p: 0.934419443391483
impulseTOsigmoid_padj: 1
sigmoidTOconst_p: 0.809685268421066
sigmoidTOconst_padj: 0.809685268421066
isTransient: FALSE
isMonotonous: FALSE
allZero: FALSE
From the figure above, it seems clear that an impulse model would fit better than a sigmoid, and a sigmoid should definitely fit better than a constant model. However, these p values seem to indicate otherwise, and I'm confused as to why this is the case. Any help or advice would be appreciated!