This is my example of dataframe (this is just a sample for testing the formulas):
dhBMEC_1 dhBMEC_2 dhBMEC_3 cryo.dhBMEC_1 cryo.dhBMEC_2 cryo.dhBMEC_3 iPSC_1 iPSC_2 iPSC_3 653635 1217 689 1089 1200 1372 1099 729 661 657 102466751 16 5 16 8 24 25 1 1 4 729737 1281 1187 1188 1482 1379 1591 3056 2799 2268
I'm doing the calculations BY HAND of the 3 steps of the DESeq2:
- Estimate Size Factors
- Estimate Dispersions
- Negative Binomial GLM fitting and Wald statistics
For first step, I was using this page: https://github.com/hbctraining/DGE_workshop_salmon/blob/master/lessons/02_DGE_count_normalization.md Using that link, i was able to make the stimation of the size factors in only 4 steps.
That's what I need right now. An easy step by step of the algorithm. The problem is, I only have the information for the step 1 but now I need to make the same for the dispersion calculation and also is the hardest step. Could anyone help me with this?
The objective is to complete this formula:
I make a list of the Inputs that I need based in the previous ecuation (i don't know if it's ok). I put a ✔ on the items that I have now:
normalized counts ✔, dispersion ✔, vector de counts ✔, estimated coefficient vector ✘, matrix model X ✘ (or ✔, but idk what it is yet)
I'm extracting all the formulas from this links:
- https://genomebiology.biomedcentral.com/track/pdf/10.1186/gb-2010-11-10-r106.pdf
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4302049/pdf/13059_2014_Article_550.pdf
- https://pdfs.semanticscholar.org/1a80/c9d6b8e918ccca0044a8f524c1bfb0295d8f.pdf?_ga=2.9250997.1440590616.1629921245-481614050.1629921245
- http://bioconductor.riken.jp/packages/3.10/bioc/vignettes/DESeq2/inst/doc/DESeq2.html

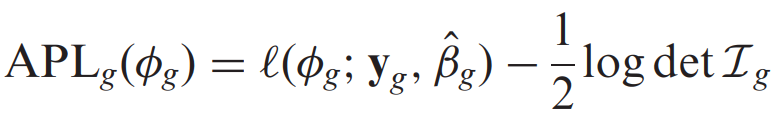

In the third paper I thought they used that calculation because it was named in the dispersion section in the Deseq2 manual and also in the second paper but it seems that I did not interpret the information correctly. I would check that.
I don't think is unrealistic, I only need the correct explanation. I already make the first step in order to complete de algorithm and maked progress in the second step of three. Everyone can spend their time telling me that I can't do it or they can use it to explain how to do it.
Anyway I appreciate your answer, Gordon, it helped me to identify that I got confused with the formula that I shared and that is really helpful to me.
Step 1 has been explained here.
However, I am also seeking to break down Steps 2 and 3 further to understand how the data is being transformed but because of limited statistical knowledge, I barely understand them theoretically only. Would love to get some inputs as DESeq2 step 2 and 3 are a bit knotty and variegated for starters!!
Yes, I found the step 1 easily. But no one could explain the other 2 steps in an easy way step by step (like the link you passed) and it's difficult to me to understand the mathematical formulas