Hi all, I have a question about the "interaction term".
my coldata is below.
sample condition patient patient_nested status
1 S02_A Pre S02 1 NR
2 S02_B Post S02 1 NR
3 S03_A Pre S03 2 NR
4 S03_B Post S03 2 NR
5 S04_A Pre S04 3 NR
6 S04_B Post S04 3 NR
7 S05_A Pre S05 4 NR
8 S05_B Post S05 4 NR
9 S06_A Pre S06 5 NR
10 S06_B Post S06 5 NR
11 S10_A Pre S10 1 R
12 S10_B Post S10 1 R
13 S11_A Pre S11 6 NR
14 S11_B Post S11 6 NR
15 S12_A Pre S12 7 NR
16 S12_B Post S12 7 NR
17 S13_A Pre S13 8 R
18 S13_B Post S13 8 R
19 S14_A Pre S14 9 NR
20 S14_B Post S14 9 NR
21 S15_A Pre S15 10 NR
22 S15_B Post S15 10 NR
23 S16_A Pre S16 11 NR
24 S16_B Post S16 11 NR
25 S17_A Pre S17 2 R
26 S17_B Post S17 2 R
In this design, I wanted to see the different effects of treatment in conditions, so I used the interaction term.
dds <- DESeqDataSetFromMatrix(countData=met,
colData=coldata,
design=~ patient_nested + condition + status + condition:status)
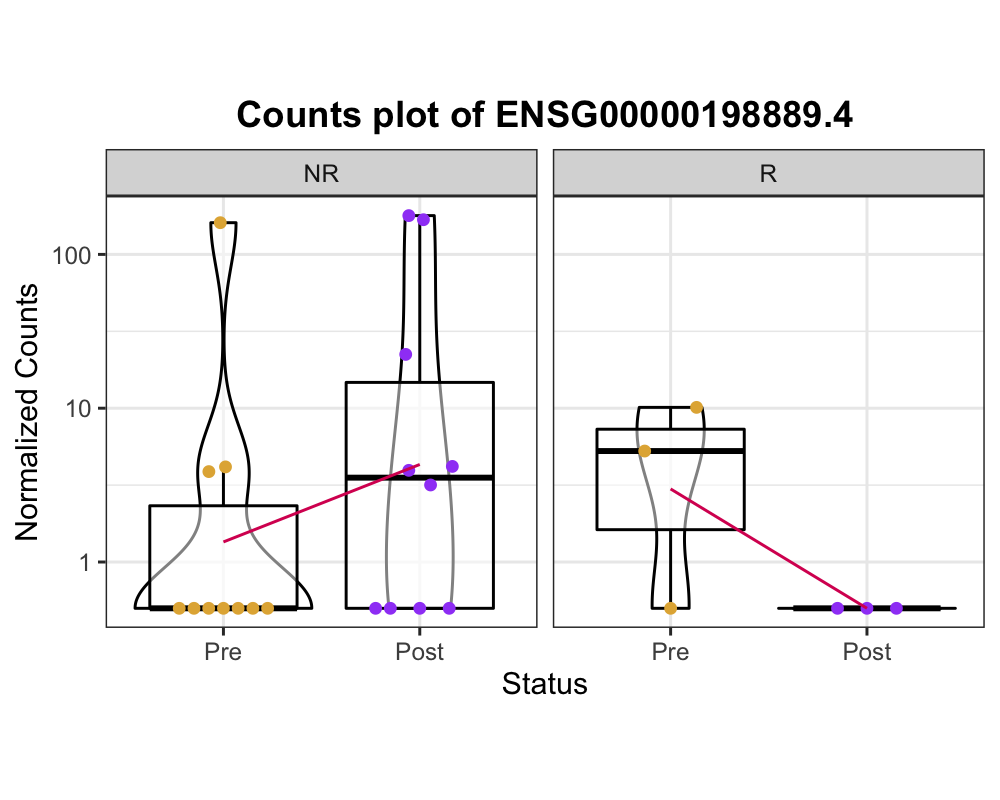
I expected the results of the different effects of treatment in conditions like below image. The expression of ENSG00000198889.4 is increased in NR and decreased in R like below image.
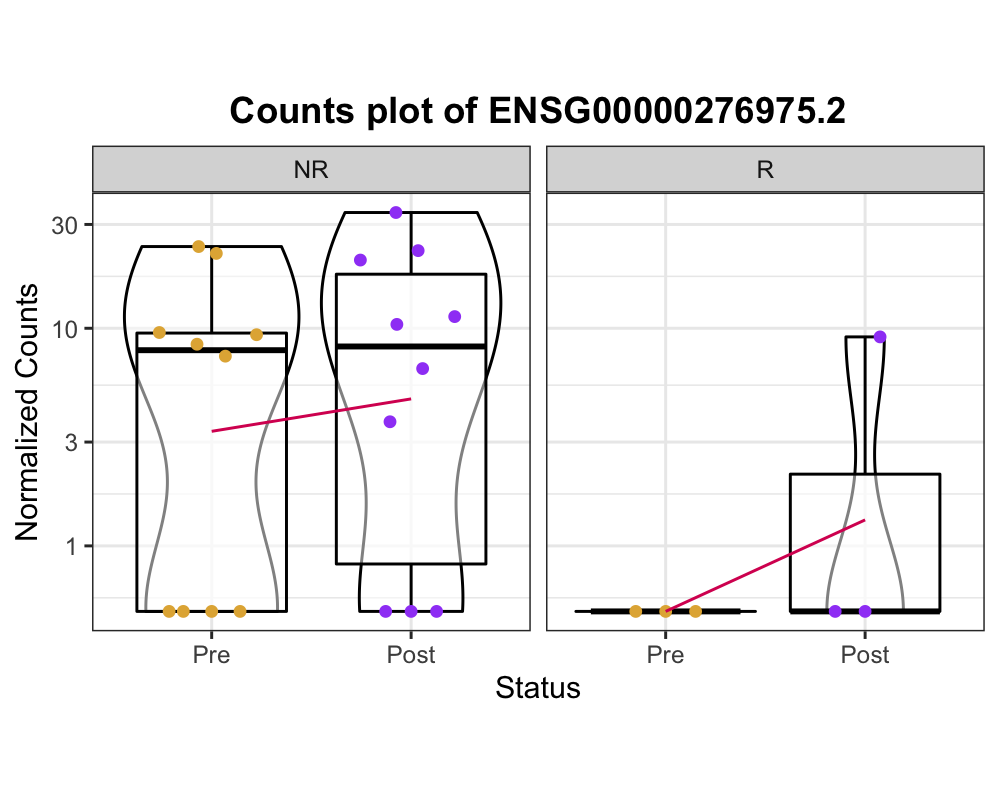
but, some of the results is not look like expected results.. see below image please.
The expression of ENSG00000276975.2 is elevated in both NR and R! It is not my expected results.
I think my design of DEseq2 is right, but I probably got this result because it was wrong.
Could you tell me why I got these result and how can I corret this problem?
Thanks!


Thanks for replying my question! But, could you explain in more detail what you said, please? I'm sorry bothering you.
If these terms are unfamiliar, I strongly recommend discussing your analysis with a statistician or someone familiar with linear models. You want to make sure to properly interpret the results.