Hello,
Crossposted https://www.biostars.org/p/9492478/
I am trying to do a differential ATACseq analysis with DiffBind3, and need to build a design that DESeq2 will accept. I have tried several relevant models, but I have been unsuccessful in achieving the contrasts I desire, without DESeq2 throwing a "Model matrix not full rank" error.
Any help with generating a correct linear model would be greatly appreciated.
I have 2 cell lines (sensitive and resistant), with 16 samples (2 replicates each= so 32 samples) at 4 times points (S1 (1hr), 12hr and 24hr) and 4 treatments (X, Y, Z). Each cell line has a baseline and each time point has a control. I included a factor column while reading into DiffBind3 using the dba command, to be able to nest samples, as outlined in the DESeq2 vignette.
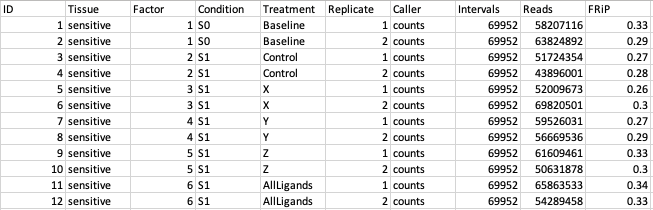
Shown here is a mock up of dba.show(db.count)[1:12]
Given that Diffbind3 subracts the controls peaks (I am guessing for each time point), my goal is to compare each treatment to baseline at every time point for both cell lines. The model I tried to build was ~Replicate+ Condition+ Condition:Factor+Treatment (for each cell line).
Any pointers or help to generate a correct linear model will be greatly appreciated.
Thank you in advance, Kavi