Greetings.
We were interested in determining unique genes in the lower canopy of legumes. We had an RNA-Seq data of Cowpea and Soybean. I am trying to analyze the Cowpea data at a SINGLE time point (flowering stage).
We had three canopy levels (Top, Middle, Low) as defined by light intensity, with five replicates for each level.
My coldata was generated as follows:
colData <- data.frame(canopy=factor(rep(c("Top","Middle","Low"),each=5)))
Some of the codes that I used were as follows:
rownames(colData) <- paste(colData$canopy, 1:5, sep = ".")
dds <- DESeqDataSetFromTximport(tx.all, colData=colData, design = ~ canopy)
colData(dds)$canopy<-relevel(colData(dds)$canopy, ref = "Top")
vsd <- vst(dds, blind=FALSE)
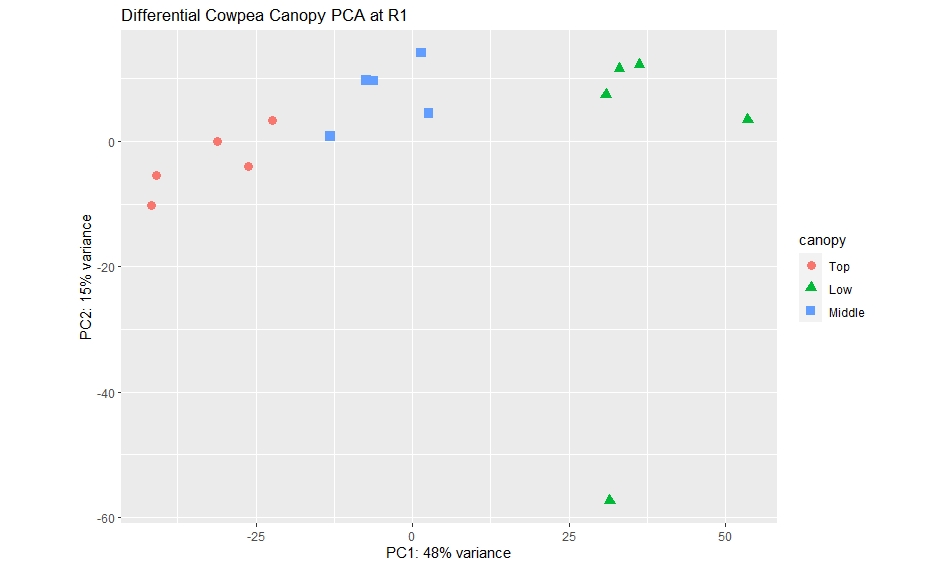
I ran the customized PCA with the "Low" canopy at the end of my colData and I got this PCA plot:
p<-(plotPCA(vsd, intgroup=c("canopy"), returnData=TRUE))
percentVar <- round(100 * attr(p, "percentVar"))
x11(10,6)
ggplot(p, aes(PC1, PC2,color=canopy, shape=canopy)) + geom_point(size=3) +
xlab(paste0("PC1: ",percentVar[1],"% variance")) +
ylab(paste0("PC2: ",percentVar[2],"% variance")) +
coord_fixed() +
ggtitle("Differential Cowpea Canopy PCA at R1")
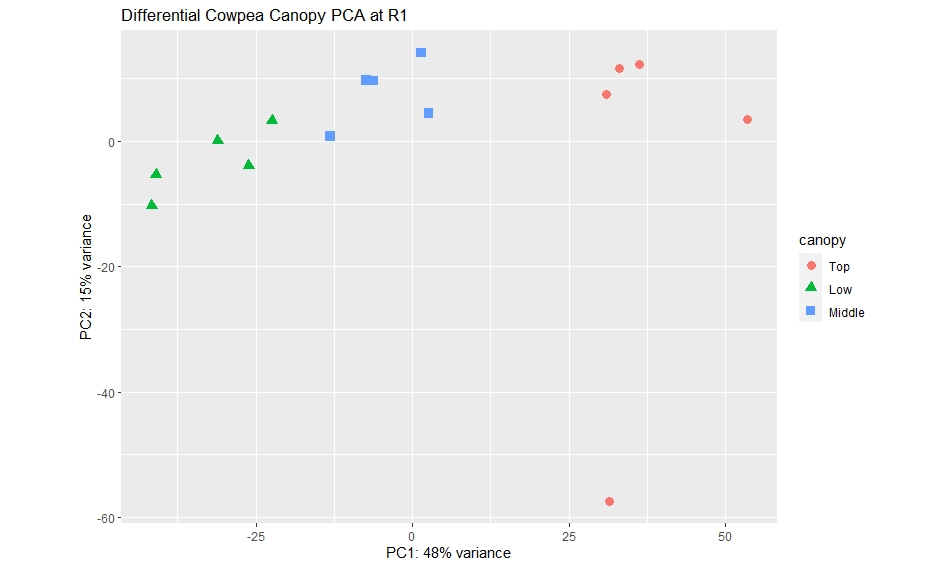
When I tried to rearranged my colData in a way that the "Top" canopy was listed last, I got a different PCA Plot as follows:
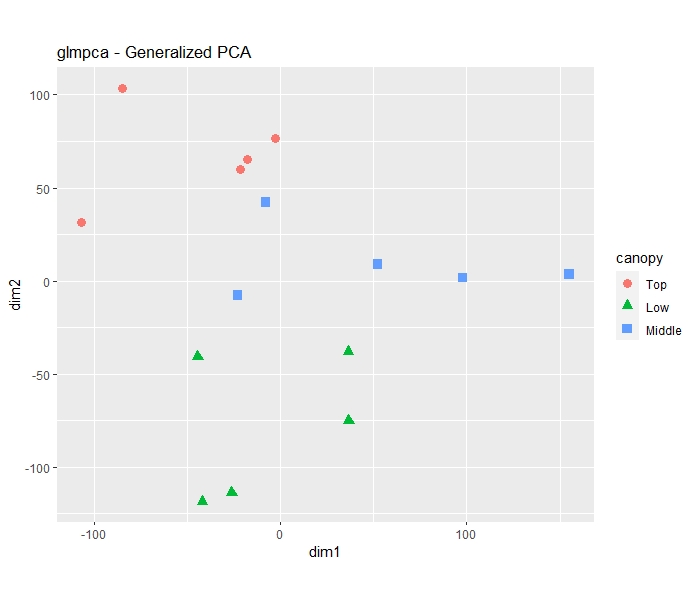
I also ran a Generalized PCA to see how it is.
library("glmpca")
gpca <- glmpca(counts(dds), L=2)
gpca.dat <- gpca$factors
gpca.dat$canopy <- dds$canopy
gpca.dat$canopy <- dds$canopy
x11(20,20)
ggplot(gpca.dat, aes(x = dim1, y = dim2, color = canopy, shape = canopy)) +
geom_point(size =3) + coord_fixed() + ggtitle("glmpca - Generalized PCA")
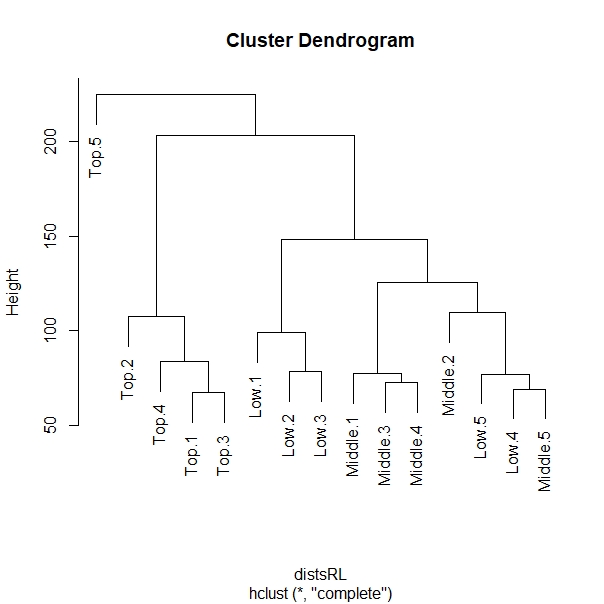
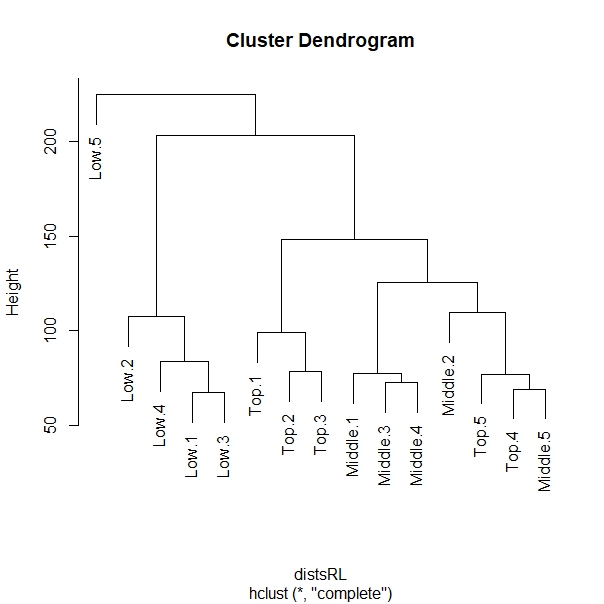
My question is that, the last variable on my colData (be it from Top or Low canopy) is showing as the root of the PCA. And it seems like an outlier based on the PCA plot and on the cluster dendrogram.
I tried removing the last variable on my colData be it from Top or Low canopy sample but it is showing the same trend, the last sample is showing like an outlier.
sessionInfo( )
R version 4.1.0 (2021-05-18)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252 LC_MONETARY=English_United States.1252
[3] LC_NUMERIC=C LC_TIME=English_United States.1252
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] WGCNA_1.70-3 fastcluster_1.2.3 dynamicTreeCut_1.63-1 dplyr_1.0.7 glmpca_0.2.0
[6] readr_2.0.1 tximport_1.21.1 genefilter_1.75.0 pheatmap_1.0.12 RColorBrewer_1.1-2
[11] ggplot2_3.3.5 gplots_3.1.1 DESeq2_1.33.4 SummarizedExperiment_1.23.1 Biobase_2.53.0
[16] MatrixGenerics_1.5.3 matrixStats_0.60.0 GenomicRanges_1.45.0 GenomeInfoDb_1.29.3 IRanges_2.27.2
[21] S4Vectors_0.31.1 BiocGenerics_0.39.2
loaded via a namespace (and not attached):
[1] colorspace_2.0-2 ellipsis_0.3.2 htmlTable_2.3.0 XVector_0.33.0 base64enc_0.1-3 rstudioapi_0.13
[7] farver_2.1.0 bit64_4.0.5 AnnotationDbi_1.56.2 fansi_0.5.0 mvtnorm_1.1-3 apeglm_1.16.0
[13] codetools_0.2-18 splines_4.1.0 doParallel_1.0.16 impute_1.68.0 cachem_1.0.5 knitr_1.36
[19] geneplotter_1.72.0 Formula_1.2-4 jsonlite_1.7.2 annotate_1.72.0 cluster_2.1.2 ashr_2.2-47
[25] GO.db_3.14.0 png_0.1-7 BiocManager_1.30.16 compiler_4.1.0 httr_1.4.2 backports_1.3.0
[31] assertthat_0.2.1 Matrix_1.3-4 fastmap_1.1.0 htmltools_0.5.2 tools_4.1.0 coda_0.19-4
[37] gtable_0.3.0 glue_1.4.2 GenomeInfoDbData_1.2.7 Rcpp_1.0.7 bbmle_1.0.24 vctrs_0.3.8
[43] Biostrings_2.61.2 preprocessCore_1.56.0 iterators_1.0.13 stringr_1.4.0 xfun_0.27 lifecycle_1.0.1
[49] irlba_2.3.3 gtools_3.9.2 XML_3.99-0.7 zlibbioc_1.39.0 MASS_7.3-54 scales_1.1.1
[55] vroom_1.5.4 hms_1.1.1 parallel_4.1.0 gridExtra_2.3 memoise_2.0.0 emdbook_1.3.12
[61] rpart_4.1-15 bdsmatrix_1.3-4 stringi_1.7.5 latticeExtra_0.6-29 RSQLite_2.2.7 SQUAREM_2021.1
[67] foreach_1.5.1 checkmate_2.0.0 caTools_1.18.2 BiocParallel_1.27.3 truncnorm_1.0-8 rlang_0.4.11
[73] pkgconfig_2.0.3 bitops_1.0-7 lattice_0.20-44 invgamma_1.1 purrr_0.3.4 htmlwidgets_1.5.4
[79] labeling_0.4.2 bit_4.0.4 tidyselect_1.1.1 plyr_1.8.6 magrittr_2.0.1 R6_2.5.1
[85] generics_0.1.1 Hmisc_4.6-0 DelayedArray_0.19.1 DBI_1.1.1 foreign_0.8-81 pillar_1.6.4
[91] withr_2.4.2 nnet_7.3-16 survival_3.2-11 KEGGREST_1.34.0 RCurl_1.98-1.4 mixsqp_0.3-43
[97] tibble_3.1.3 crayon_1.4.2 KernSmooth_2.23-20 utf8_1.2.2 tzdb_0.1.2 jpeg_0.1-9
[103] locfit_1.5-9.4 grid_4.1.0 data.table_1.14.2 blob_1.2.2 digest_0.6.28 xtable_1.8-4
[109] numDeriv_2016.8-1.1 munsell_0.5.0