Entering edit mode
I have 20 datasets from different parts and developmental stages of the fruit (seed, flesh fruit, peel).
A total of 205,482 genes were retained after filtering genes (with TPM values) with coefficient of variation (cv) <1. As the number is still big, further filtering was done to obtain ONLY 70% most variable genes using the R code below. This ended up with 61,645 genes to proceed with WGCNA .
mData0$variance = apply(mData0, 1, var)
mData2 = mData0[mData0$variance >= quantile(mData0$variance, c(.70)), ]
- Is my strategy in data cleaning and pre-processing correct?
- This data suitable for unsigned or signed network?
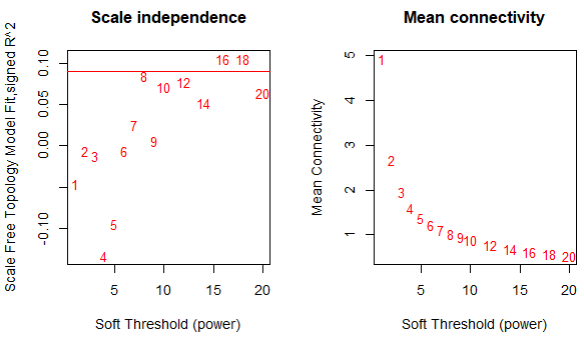
- I'm planning to perform automatic network construction function blockwise. However, may I know how to choose soft threshold value for WGCNA analysis based on the figure below?