Entering edit mode
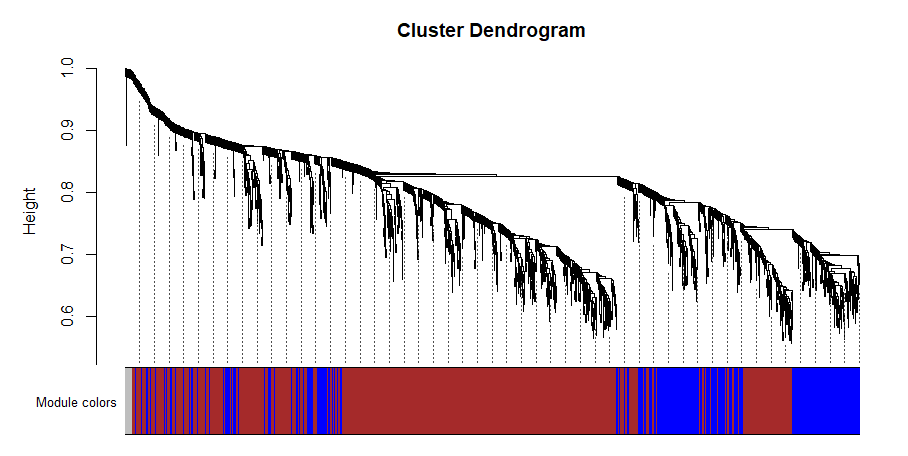
I am trying to plot my gene dendrogram while following the online tutorials for WGCNA. When using the function "table(bwnet$colors)" it shows that there should be 24 modules for my data. When I continue running the code for plotting the dendrogram (code below) I somehow keep getting a plot that looks like this. It seems to me that this is not consistent with there being 24 modules.
bwnet = blockwiseModules(datExpr0, maxBlockSize = 2000,
power = 6, TOMType = "unsigned", minModuleSize = 30,
reassignThreshold = 0, mergeCutHeight = 0.25,
numericLabels = TRUE,
saveTOMs = TRUE,
saveTOMFileBase = "TOM-blockwise",
verbose = 3)
# open a graphics window
sizeGrWindow(12, 9)
# Convert labels to colors for plotting
mergedColors = labels2colors(bwnet$colors)
# Plot the dendrogram and the module colors underneath
plotDendroAndColors(bwnet$dendrograms[[1]], mergedColors[bwnet$blockGenes[[1]]],
"Module colors",
dendroLabels = FALSE, hang = 0.03,
addGuide = TRUE, guideHang = 0.05)
table(bwnet$colors)
![> 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22
1287 4001 3115 2498 1774 1706 1135 735 484 305 299 269 265 157 117 111 104 101 77 71 68 63 61
23 24
60 44][1]


Hello,
I think we have the same problem, have you figured it out?