This question relates to my previous post (Advice on designing an EdgeR object in complex experimental setup ?)
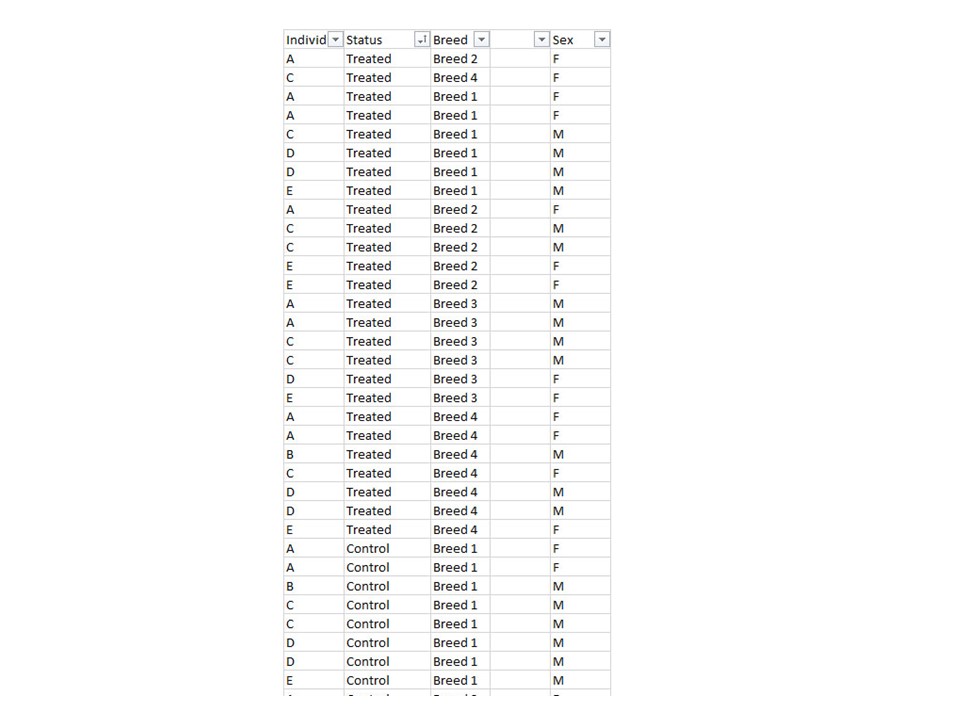
I have an experimental design like in this image
I first created a data frame for the two groups (Animals) and (Status):
Animal <- factor(paste(Sample_info$Individual, Sample_info$Breed, sep = '.'))
Status<-factor(Sample_info$Status, levels = c('Treated', 'Control'))
Then I made the 'design matrix'
design <- model.matrix(~Animal+Status)
which produced a design matrix that looks like this 
My question is why the design matrix named its last column ' StatusControl' and not 'statustreated' ? this caused it to label the control samples as 1 and treated as 0, which I assume that this told the program to consider the treated groups as a 'reference' and control group as 'comparison' group so that all my genes (which in the count was higher in treated samples than in control) appeared as downregulated ? It should be the reverse..... So I want to tell R to consider the control as a 'reference group' and I think label it as 0 (not 1). How can I do that ?
This was my DE code :
fit<- glmQLFit(y, design, robust = TRUE)
tr <- glmQLFTest(fit, coef = "StatusUnstimulated")
I would be appreciating any answer !


Thanks a lot, will try this and see.
When I answered your original question, I assumed you would use the simplest possible code
which would have given the correct result. By default, the factor levels are ordered in alphabetical order, and "Control" comes before "Treated" alphabetically.