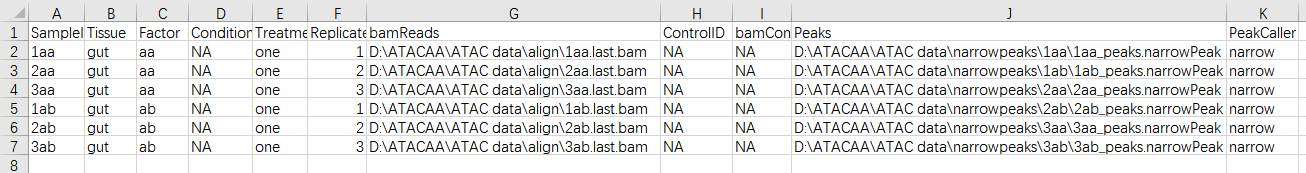
Hello,
I have ATAC sequencing data for two types of cells, each with three replicates. Get strange results by DiffBind analysis. Why is my volcano map only halfway? I don't know where I went wrong. Is there a problem with the original data or with the processing results? Can anyone answer it. here is my SampleSheet.
last.BAM was compared to the genome.Mitochondrial duplication, PCR duplication and low-quality reads were removed. Paired-ended sequencing to the same chromosome
here is my code
a <- dba(sampleSheet="diff.csv")
a <- dba.count(a,bUseSummarizeOverlaps = TRUE)
#warning:In serialize(data, node$con) :
#'package:stats' may not be available when loading
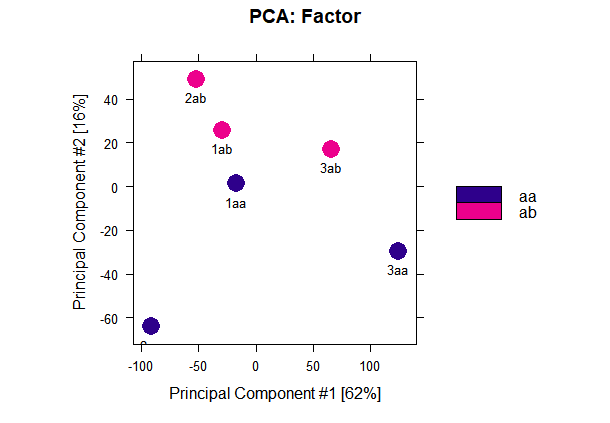
dba.plotPCA(a,attributes=DBA_FACTOR,label=DBA_ID)
#figure2
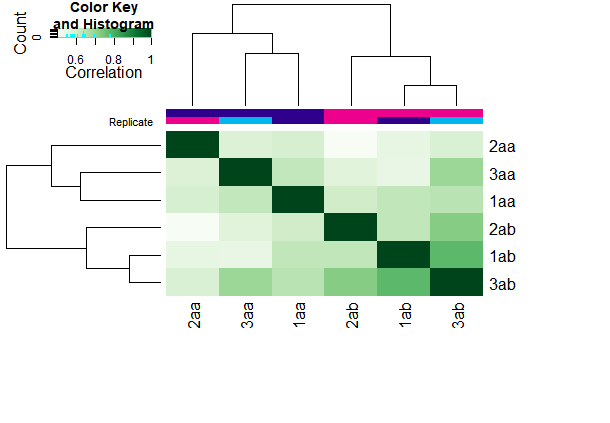
plot(a)
#figure3
a <- dba.contrast(a,categories = DBA_FACTOR,minMembers = 2)
a <- dba.analyze(a,method=DBA_DESEQ2)
#warning: solve(): system is singular (rcond: 3.89123e-18); attempting approx solution
dba.show(a,bContrasts = T)
# Factor Group Samples Group2 Samples2 DB.DESeq2
# 1 Factor ab 3 aa 3 8
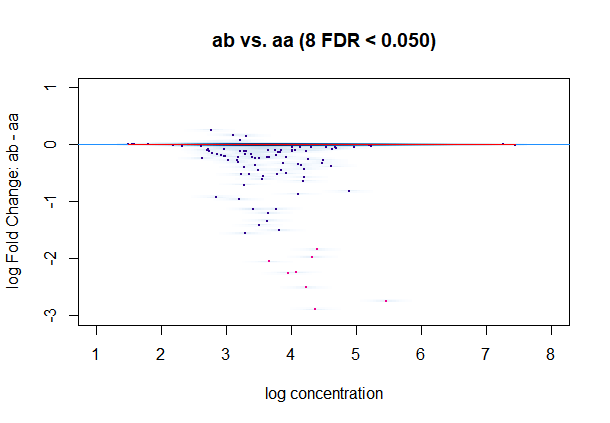
dba.plotMA(a)
#warning:In KernSmooth::bkde2D(x, bandwidth = bandwidth, gridsize = nbin, :Binning grid too coarse for current (small) bandwidth: consider increasing 'gridsize'
#figure4
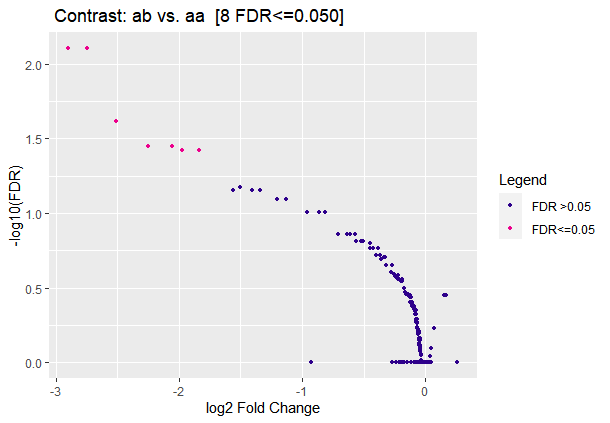
#This picture is weird.
dba.plotVolcano(a)
#figure5
sessionInfo( )