I am doing a GSEA analysis with clusterprofiler, but I am getting an error in ridgeplot. This is my code. I get an error even if I set pvalueCutoff to 1. How can I solve the problem? Thanks.
,,,df = read.csv("T00.csv", header=TRUE)
,,,original_gene_list_0_2 <- df_0$logFC
,,,names(original_gene_list_0_2) <- df_0$X
,,,head(original_gene_list_0_2)
,,,gene_list_0_2<-na.omit(original_gene_list_0_2)
,,,gene_list_0_2 = sort(gene_list_0_2, decreasing = TRUE)
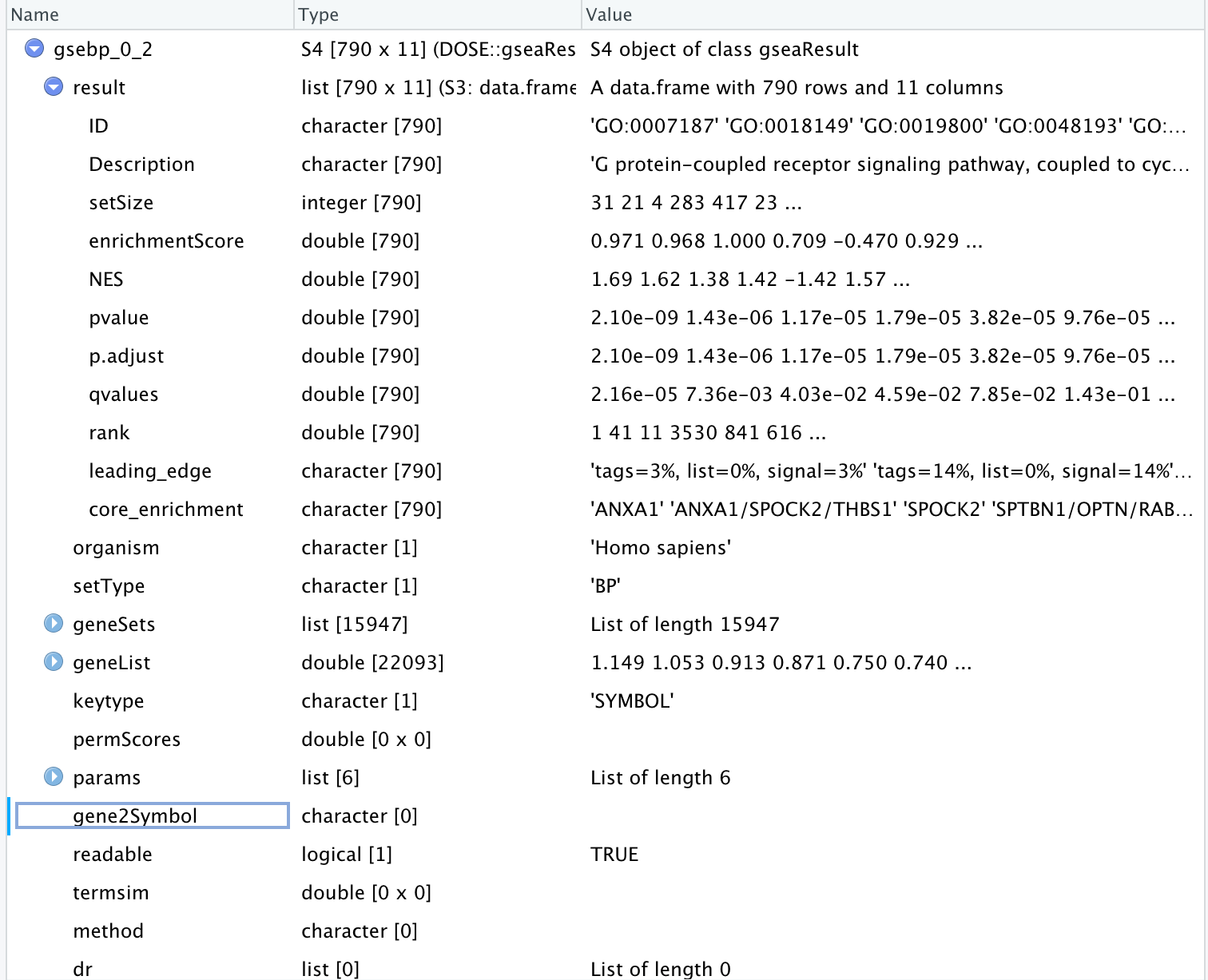
,,,gsebp_0_2 <- gseGO(geneList=gene_list_0_2, ont="BP", keyType = "SYMBOL",
minGSSize = 3,
maxGSSize = 800,
pvalueCutoff = 0.05,
verbose = TRUE,
nPermSimple = 10000,
OrgDb =org.Hs.eg.db,
pAdjustMethod = "none")
,,,ridgeplot(gsebp_0_2,showCategory = 8)
,,,Error in ans[ypos] <- rep(yes, length.out = len)[ypos] :
replacement has length zero
In addition: Warning message:
In rep(yes, length.out = len) : 'x' is NULL so the result will be NULL
sessionInfo( )
setting value version R version 4.2.1 (2022-06-23) os macOS Monterey 12.4 system x86_64, darwin17.0 ui RStudio language (EN) collate en_US.UTF-8 ctype en_US.UTF-8 tz Asia/Tokyo date 2022-07-22 rstudio 2022.07.0+548 Spotted Wakerobin (desktop) pandoc NA
─ Packages package version date (UTC) lib source abind 1.4-5 2016-07-21 1 CRAN (R 4.2.0) AnnotationDbi 1.58.0 2022-04-26 1 Bioconductor AnnotationHub 3.4.0 2022-04-26 1 Bioconductor ape 5.6-2 2022-03-02 1 CRAN (R 4.2.0) aplot 0.1.6 2022-06-03 1 CRAN (R 4.2.0) assertthat 0.2.1 2019-03-21 1 CRAN (R 4.2.0) babelgene 22.3 2022-03-30 1 CRAN (R 4.2.0) Biobase 2.56.0 2022-04-26 1 Bioconductor BiocFileCache 2.4.0 2022-04-26 1 Bioconductor BiocGenerics 0.42.0 2022-04-26 1 Bioconductor BiocManager 1.30.18 2022-05-18 1 CRAN (R 4.2.0) BiocParallel 1.30.3 2022-06-05 1 Bioconductor BiocVersion 3.15.2 2022-03-29 1 Bioconductor Biostrings 2.64.0 2022-04-26 1 Bioconductor bit 4.0.4 2020-08-04 1 CRAN (R 4.2.0) bit64 4.0.5 2020-08-30 1 CRAN (R 4.2.0) bitops 1.0-7 2021-04-24 1 CRAN (R 4.2.0) blob 1.2.3 2022-04-10 1 CRAN (R 4.2.0) cachem 1.0.6 2021-08-19 1 CRAN (R 4.2.0) callr 3.7.1 2022-07-13 1 CRAN (R 4.2.0) cli 3.3.0 2022-04-25 1 CRAN (R 4.2.0) cluster 2.1.3 2022-03-28 1 CRAN (R 4.2.1) clusterProfiler 4.5.1.902 2022-07-21 1 Github (GuangchuangYu/clusterProfiler@48ad00f) codetools 0.2-18 2020-11-04 1 CRAN (R 4.2.1) colorspace 2.0-3 2022-02-21 1 CRAN (R 4.2.0) cowplot 1.1.1 2020-12-30 1 CRAN (R 4.2.0) crayon 1.5.1 2022-03-26 1 CRAN (R 4.2.0) curl 4.3.2 2021-06-23 1 CRAN (R 4.2.0) data.table 1.14.2 2021-09-27 1 CRAN (R 4.2.0) DBI 1.1.3 2022-06-18 1 CRAN (R 4.2.0) dbplyr 2.2.1 2022-06-27 1 CRAN (R 4.2.0) deldir 1.0-6 2021-10-23 1 CRAN (R 4.2.0) devtools 2.4.4 2022-07-20 1 CRAN (R 4.2.1) digest 0.6.29 2021-12-01 1 CRAN (R 4.2.0) DO.db 2.9 2022-07-19 1 Bioconductor DOSE 3.23.2 2022-07-21 1 Github (GuangchuangYu/DOSE@b42425a) downloader 0.4 2015-07-09 1 CRAN (R 4.2.0) dplyr 1.0.9 2022-04-28 1 CRAN (R 4.2.0) ellipsis 0.3.2 2021-04-29 1 CRAN (R 4.2.0) enrichplot 1.17.0.992 2022-07-21 1 Github (YuLab-SMU/enrichplot@a546701) europepmc 0.4.1 2021-09-02 1 CRAN (R 4.2.0) fansi 1.0.3 2022-03-24 1 CRAN (R 4.2.0) farver 2.1.1 2022-07-06 1 CRAN (R 4.2.0) fastmap 1.1.0 2021-01-25 1 CRAN (R 4.2.0) fastmatch 1.1-3 2021-07-23 1 CRAN (R 4.2.0) fgsea 1.22.0 2022-04-26 1 Bioconductor filelock 1.0.2 2018-10-05 1 CRAN (R 4.2.0) fitdistrplus 1.1-8 2022-03-10 1 CRAN (R 4.2.0) fs 1.5.2 2021-12-08 1 CRAN (R 4.2.0) future 1.26.1 2022-05-27 1 CRAN (R 4.2.0) future.apply 1.9.0 2022-04-25 1 CRAN (R 4.2.0) generics 0.1.3 2022-07-05 1 CRAN (R 4.2.0) GenomeInfoDb 1.32.2 2022-05-15 1 Bioconductor GenomeInfoDbData 1.2.8 2022-07-19 1 Bioconductor ggforce 0.3.3 2021-03-05 1 CRAN (R 4.2.0) ggfun 0.0.6 2022-04-01 1 CRAN (R 4.2.0) ggnewscale 0.4.7.9000 2022-07-20 1 Github (eliocamp/ggnewscale@298375e) ggplot2 3.3.6.9000 2022-07-21 1 Github (tidyverse/ggplot2@a979ffd) ggplotify 0.1.0 2021-09-02 1 CRAN (R 4.2.0) ggraph 2.0.5 2021-02-23 1 CRAN (R 4.2.0) ggrepel 0.9.1 2021-01-15 1 CRAN (R 4.2.0) ggridges 0.5.3 2021-01-08 1 CRAN (R 4.2.0) ggtree 3.4.1 2022-07-14 1 Bioconductor ggupset 0.3.0 2020-05-05 1 CRAN (R 4.2.0) globals 0.15.1 2022-06-24 1 CRAN (R 4.2.0) glue 1.6.2 2022-02-24 1 CRAN (R 4.2.0) GO.db 3.15.0 2022-07-19 1 Bioconductor goftest 1.2-3 2021-10-07 1 CRAN (R 4.2.0) GOSemSim 2.22.0 2022-04-26 1 Bioconductor graph 1.74.0 2022-04-26 1 Bioconductor graphlayouts 0.8.0 2022-01-03 1 CRAN (R 4.2.0) gridExtra 2.3 2017-09-09 1 CRAN (R 4.2.0) gridGraphics 0.5-1 2020-12-13 1 CRAN (R 4.2.0) gson 0.0.6 2022-06-07 1 CRAN (R 4.2.0) gtable 0.3.0 2019-03-25 1 CRAN (R 4.2.0) hms 1.1.1 2021-09-26 1 CRAN (R 4.2.0) htmltools 0.5.3 2022-07-18 1 CRAN (R 4.2.1) htmlwidgets 1.5.4 2021-09-08 1 CRAN (R 4.2.0) httpuv 1.6.5 2022-01-05 1 CRAN (R 4.2.0) httr 1.4.3 2022-05-04 1 CRAN (R 4.2.0) ica 1.0-3 2022-07-08 1 CRAN (R 4.2.0) igraph 1.3.4 2022-07-19 1 CRAN (R 4.2.0) interactiveDisplayBase 1.34.0 2022-04-26 1 Bioconductor IRanges 2.30.0 2022-04-26 1 Bioconductor irlba 2.3.5 2021-12-06 1 CRAN (R 4.2.0) jsonlite 1.8.0 2022-02-22 1 CRAN (R 4.2.0) KEGGREST 1.36.3 2022-07-14 1 Bioconductor KernSmooth 2.23-20 2021-05-03 1 CRAN (R 4.2.1) labeling 0.4.2 2020-10-20 1 CRAN (R 4.2.0) later 1.3.0 2021-08-18 1 CRAN (R 4.2.0) lattice 0.20-45 2021-09-22 1 CRAN (R 4.2.1) lazyeval 0.2.2 2019-03-15 1 CRAN (R 4.2.0) leiden 0.4.2 2022-05-09 1 CRAN (R 4.2.0) lifecycle 1.0.1 2021-09-24 1 CRAN (R 4.2.0) listenv 0.8.0 2019-12-05 1 CRAN (R 4.2.0) lmtest 0.9-40 2022-03-21 1 CRAN (R 4.2.0) magrittr 2.0.3 2022-03-30 1 CRAN (R 4.2.0) MASS 7.3-58 2022-07-14 1 CRAN (R 4.2.0) Matrix 1.4-1 2022-03-23 1 CRAN (R 4.2.1) matrixStats 0.62.0 2022-04-19 1 CRAN (R 4.2.0) memoise 2.0.1 2021-11-26 1 CRAN (R 4.2.0) mgcv 1.8-40 2022-03-29 1 CRAN (R 4.2.1) mime 0.12 2021-09-28 1 CRAN (R 4.2.0) miniUI 0.1.1.1 2018-05-18 1 CRAN (R 4.2.0) msigdbr 7.5.1 2022-03-30 1 CRAN (R 4.2.0) munsell 0.5.0 2018-06-12 1 CRAN (R 4.2.0) nlme 3.1-158 2022-06-15 1 CRAN (R 4.2.0) org.Hs.eg.db 3.15.0 2022-07-19 1 Bioconductor parallelly 1.32.1 2022-07-21 1 CRAN (R 4.2.1) patchwork 1.1.1 2020-12-17 1 CRAN (R 4.2.0) pbapply 1.5-0 2021-09-16 1 CRAN (R 4.2.0) pillar 1.8.0 2022-07-18 1 CRAN (R 4.2.1) pkgbuild 1.3.1 2021-12-20 1 CRAN (R 4.2.0) pkgconfig 2.0.3 2019-09-22 1 CRAN (R 4.2.0) pkgload 1.3.0 2022-06-27 1 CRAN (R 4.2.0) plotly 4.10.0 2021-10-09 1 CRAN (R 4.2.0) plyr 1.8.7 2022-03-24 1 CRAN (R 4.2.0) png 0.1-7 2013-12-03 1 CRAN (R 4.2.0) polyclip 1.10-0 2019-03-14 1 CRAN (R 4.2.0) presto 1.0.0 2022-07-19 1 Github (immunogenomics/presto@052085d) prettyunits 1.1.1 2020-01-24 1 CRAN (R 4.2.0) processx 3.7.0 2022-07-07 1 CRAN (R 4.2.0) profvis 0.3.7 2020-11-02 1 CRAN (R 4.2.0) progress 1.2.2 2019-05-16 1 CRAN (R 4.2.0) progressr 0.10.1 2022-06-03 1 CRAN (R 4.2.0) promises 1.2.0.1 2021-02-11 1 CRAN (R 4.2.0) ps 1.7.1 2022-06-18 1 CRAN (R 4.2.0) purrr 0.3.4 2020-04-17 1 CRAN (R 4.2.0) qvalue 2.28.0 2022-04-26 1 Bioconductor R6 2.5.1 2021-08-19 1 CRAN (R 4.2.0) RANN 2.6.1 2019-01-08 1 CRAN (R 4.2.0) rappdirs 0.3.3 2021-01-31 1 CRAN (R 4.2.0) RColorBrewer 1.1-3 2022-04-03 1 CRAN (R 4.2.0) Rcpp 1.0.9 2022-07-08 1 CRAN (R 4.2.0) RcppAnnoy 0.0.19 2021-07-30 1 CRAN (R 4.2.0) RCurl 1.98-1.7 2022-06-09 1 CRAN (R 4.2.0) remotes 2.4.2 2021-11-30 1 CRAN (R 4.2.0) reshape2 1.4.4 2020-04-09 1 CRAN (R 4.2.0) reticulate 1.25 2022-05-11 1 CRAN (R 4.2.0) rgeos 0.5-9 2021-12-15 1 CRAN (R 4.2.0) Rgraphviz 2.40.0 2022-04-26 1 Bioconductor rlang 1.0.4 2022-07-12 1 CRAN (R 4.2.0) ROCR 1.0-11 2020-05-02 1 CRAN (R 4.2.0) rpart 4.1.16 2022-01-24 1 CRAN (R 4.2.1) RSQLite 2.2.15 2022-07-17 1 CRAN (R 4.2.1) rstudioapi 0.13 2020-11-12 1 CRAN (R 4.2.0) Rtsne 0.16 2022-04-17 1 CRAN (R 4.2.0) S4Vectors 0.34.0 2022-04-26 1 Bioconductor scales 1.2.0 2022-04-13 1 CRAN (R 4.2.0) scattermore 0.8 2022-02-14 1 CRAN (R 4.2.0) scatterpie 0.1.7 2021-08-20 1 CRAN (R 4.2.0) sctransform 0.3.3 2022-01-13 1 CRAN (R 4.2.0) sessioninfo 1.2.2 2021-12-06 1 CRAN (R 4.2.0) Seurat 4.1.1 2022-05-02 1 CRAN (R 4.2.0) SeuratObject 4.1.0 2022-05-01 1 CRAN (R 4.2.0) shadowtext 0.1.2 2022-04-22 1 CRAN (R 4.2.0) shiny 1.7.2 2022-07-19 1 CRAN (R 4.2.1) sp 1.5-0 2022-06-05 1 CRAN (R 4.2.0) SparseM 1.81 2021-02-18 1 CRAN (R 4.2.0) spatstat.core 2.4-4 2022-05-18 1 CRAN (R 4.2.0) spatstat.data 2.2-0 2022-04-18 1 CRAN (R 4.2.0) spatstat.geom 2.4-0 2022-03-29 1 CRAN (R 4.2.0) spatstat.random 2.2-0 2022-03-30 1 CRAN (R 4.2.0) spatstat.sparse 2.1-1 2022-04-18 1 CRAN (R 4.2.0) spatstat.utils 2.3-1 2022-05-06 1 CRAN (R 4.2.0) stringi 1.7.8 2022-07-11 1 CRAN (R 4.2.0) stringr 1.4.0 2019-02-10 1 CRAN (R 4.2.0) survival 3.3-1 2022-03-03 1 CRAN (R 4.2.1) tensor 1.5 2012-05-05 1 CRAN (R 4.2.0) tibble 3.1.7 2022-05-03 1 CRAN (R 4.2.0) tidygraph 1.2.1 2022-04-05 1 CRAN (R 4.2.0) tidyr 1.2.0 2022-02-01 1 CRAN (R 4.2.0) tidyselect 1.1.2 2022-02-21 1 CRAN (R 4.2.0) tidytree 0.3.9 2022-03-04 1 CRAN (R 4.2.0) topGO 2.48.0 2022-04-26 1 Bioconductor treeio 1.20.1 2022-07-14 1 Bioconductor triebeard 0.3.0 2016-08-04 1 CRAN (R 4.2.0) tweenr 1.0.2 2021-03-23 1 CRAN (R 4.2.0) urlchecker 1.0.1 2021-11-30 1 CRAN (R 4.2.0) urltools 1.7.3 2019-04-14 1 CRAN (R 4.2.0) usethis * 2.1.6 2022-05-25 1 CRAN (R 4.2.0) utf8 1.2.2 2021-07-24 1 CRAN (R 4.2.0) uwot 0.1.11 2021-12-02 1 CRAN (R 4.2.0) vctrs 0.4.1 2022-04-13 1 CRAN (R 4.2.0) viridis 0.6.2 2021-10-13 1 CRAN (R 4.2.0) viridisLite 0.4.0 2021-04-13 1 CRAN (R 4.2.0) withr 2.5.0 2022-03-03 1 CRAN (R 4.2.0) xml2 1.3.3 2021-11-30 1 CRAN (R 4.2.0) xtable 1.8-4 2019-04-21 1 CRAN (R 4.2.0) XVector 0.36.0 2022-04-26 1 Bioconductor yaml 2.3.5 2022-02-21 1 CRAN (R 4.2.0) yulab.utils 0.0.5 2022-06-30 1 CRAN (R 4.2.0) zlibbioc 1.42.0 2022-04-26 1 Bioconductor zoo 1.8-10 2022-04-15 1 CRAN (R 4.2.0)
1 /Library/Frameworks/R.framework/Versions/4.2/Resources/library


Hi Y.k, have you solved your problem? I have same error.
Because of lack of formatting, the post of the TS is difficult to read. Combined with the unavailability of the input data that was used, this question was not answered.
Having said this, the code works in my hands!
Since the input data was unavailable, I used as input the example dataset included with the library
DOSE. Moreover, I included the argumenteps=0. All other arguments are the same as used by the TS.So I suggest you first check whether the code below runs for you as well. If not, be sure to update to the latest version of all libraries. If it does, then continue with your input data. If you then run into problems, fee free to post here (with sufficient details!).
I had the same problem, which was solved when I used "EMSEMBL" or "ENTREZID" for keyType matching. I guess some duplicated SYMBOLs in our data may cause the issue.