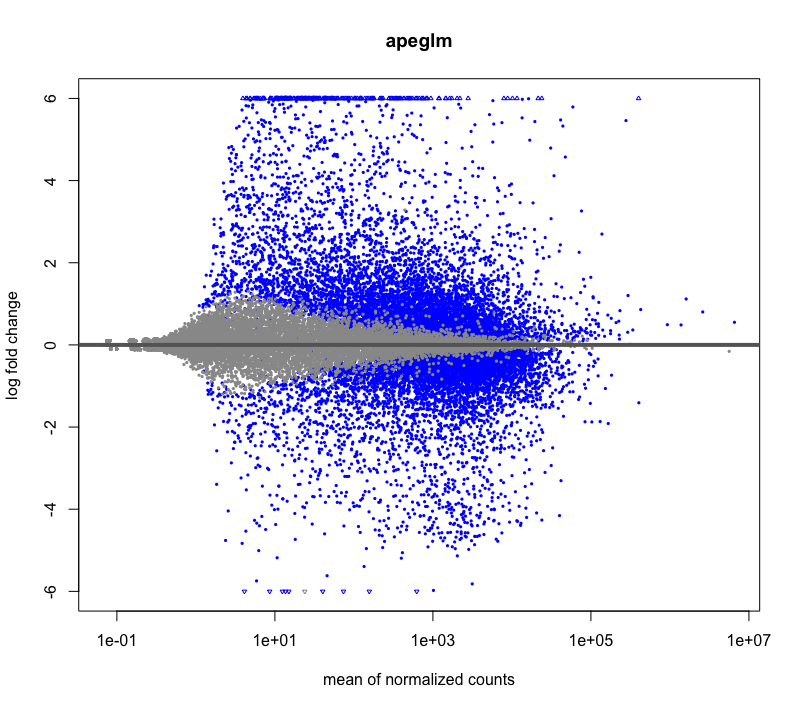
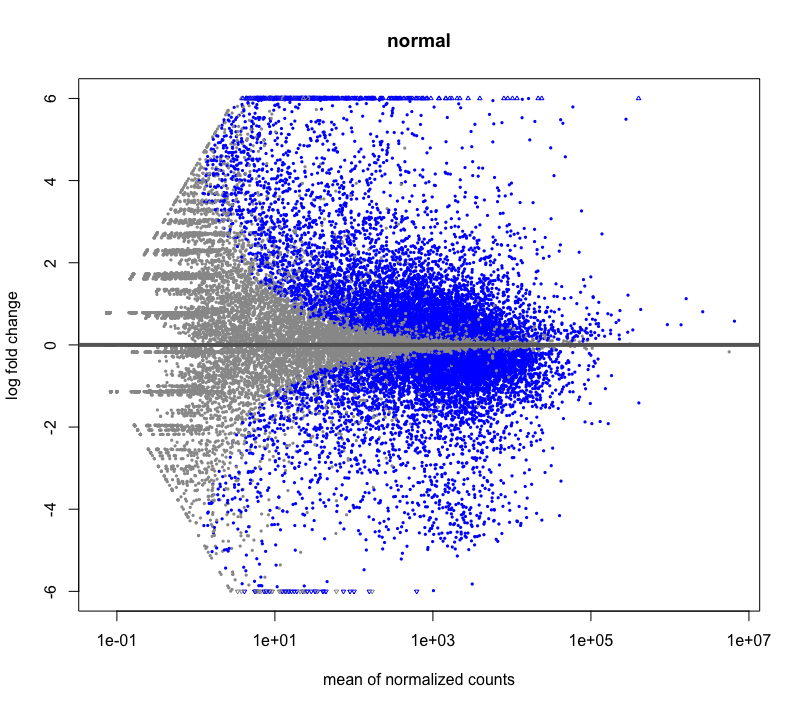
Hi, I have a question about DESeq2 LFCshrinkage: Is it possible that some genes have a slightly higher LFC after shrinkage? It happened during my RNAseq DE analysis, I have very deeply sequenced samples with large base means. I tried visualizing using MAplot check, and it looks fine. I'm mainly just curious why it could be happening.
Thank you very much!
Before shrinkage
gene_name gene_biotype baseMean log2FoldChange lfcSE stat pvalue padj
Man2b1 protein_coding 13964.1811689668 1.14901659101663 0.026320542427396 43.6547458771468 0 0
Acsl1 protein_coding 12428.1198150372 -1.19203974862738 0.0309164869284314 -38.5567658895669 0 0
Lpar1 protein_coding 10110.7963562404 -1.2202928328298 0.0304299001078297 -40.1017692633114 0 0
Plbd2 protein_coding 11931.1301365076 1.3739819601851 0.0319518259803669 43.0016726126813 0 0
Colgalt1 protein_coding 20463.1536063121 -1.41111184542647 0.0360342333779327 -39.1603126567592 0 0
Pld3 protein_coding 10409.4761223403 1.45876217014098 0.0342221971577909 42.6261985288365 0 0
Serpinh1 protein_coding 130803.363492491 -1.60180488761581 0.038333572697764 -41.7859535359521 0 0
Itm2c protein_coding 16741.2345729904 1.61533171987466 0.0363497339741802 44.4386118760058 0 0
Fam20c protein_coding 31312.9603458304 1.66573800250627 0.0356904438344024 46.6718209007153 0 0
Arl6ip5 protein_coding 6917.92975304822 1.99977697101572 0.049957690177568 40.029412166731 0 0
Ctsa protein_coding 19094.5177324866 2.01155711378371 0.0450333903613202 44.668125087723 0 0
Aplp1 protein_coding 6144.4413345581 2.03447790224768 0.0423078628847808 48.0874656275663 0 0
After shrinkage
gene_name gene_biotype baseMean log2FoldChange lfcSE pvalue padj
Man2b1 protein_coding 13964.1811689668 1.15088428333462 0.0263176482387476 0 0
Acsl1 protein_coding 12428.1198150372 -1.1904574704259 0.0309164525315082 0 0
Lpar1 protein_coding 10110.7963562404 -1.21964450839398 0.0304314704060129 0 0
Plbd2 protein_coding 11931.1301365076 1.37367794538455 0.0319515611635686 0 0
Colgalt1 protein_coding 20463.1536063121 -1.40983149622625 0.0360380229694581 0 0
Pld3 protein_coding 10409.4761223403 1.45774280936718 0.0342249709778493 0 0
Serpinh1 protein_coding 130803.363492491 -1.60093511162113 0.0383282083050976 0 0
Itm2c protein_coding 16741.2345729904 1.61350967070109 0.0363573760545177 0 0
Fam20c protein_coding 31312.9603458304 1.66751229278783 0.0356897852459903 0 0
Arl6ip5 protein_coding 6917.92975304822 1.99771245849146 0.0499730301660154 0 0
Ctsa protein_coding 19094.5177324866 2.01001220283617 0.0450386387713646 0 0
Aplp1 protein_coding 6144.4413345581 2.03381754211172 0.0423164419717999 0 0
> condsToCompare
[1] "CTRL" "24h"
res<-results(dds, contrast=c("condition", condsToCompare[2], condsToCompare[1]))
resLFC <- lfcShrink(dds, coef = "condition_24h_vs_CTRL", type="apeglm")
sessionInfo()
R version 4.0.4 (2021-02-15)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS 12.3
Matrix products: default
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] tidyr_1.2.0 edgeR_3.32.1 limma_3.46.0
[4] data.table_1.14.2 biomaRt_2.50.1 tximport_1.18.0
[7] dendsort_0.3.4 pheatmap_1.0.12 ggpubr_0.4.0
[10] openxlsx_4.2.5 EnhancedVolcano_1.8.0 PCAtools_2.2.0
[13] ggrepel_0.9.1 gplots_3.1.1 DESeq2_1.30.1
[16] SummarizedExperiment_1.20.0 Biobase_2.50.0 MatrixGenerics_1.2.1
[19] matrixStats_0.61.0 GenomicRanges_1.42.0 GenomeInfoDb_1.26.7
[22] IRanges_2.24.1 S4Vectors_0.28.1 BiocGenerics_0.36.1
[25] dplyr_1.0.8 magrittr_2.0.3 ggplot2_3.3.5
[28] RColorBrewer_1.1-3 BiocManager_1.30.16
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-3 ggsignif_0.6.3 ellipsis_0.3.2
[5] XVector_0.30.0 rstudioapi_0.13 farver_2.1.0 bit64_4.0.5
[9] mvtnorm_1.1-3 apeglm_1.12.0 AnnotationDbi_1.52.0 fansi_1.0.3
[13] xml2_1.3.3 splines_4.0.4 sparseMatrixStats_1.2.1 extrafont_0.18
[17] cachem_1.0.6 geneplotter_1.68.0 broom_0.7.12 Rttf2pt1_1.3.10
[21] annotate_1.68.0 dbplyr_2.1.1 readr_2.1.2 compiler_4.0.4
[25] httr_1.4.2 dqrng_0.3.0 backports_1.4.1 assertthat_0.2.1
[29] Matrix_1.4-1 fastmap_1.1.0 cli_3.2.0 BiocSingular_1.6.0
[33] prettyunits_1.1.1 tools_4.0.4 rsvd_1.0.5 coda_0.19-4
[37] gtable_0.3.0 glue_1.6.2 GenomeInfoDbData_1.2.4 reshape2_1.4.4
[41] rappdirs_0.3.3 maps_3.4.0 Rcpp_1.0.8.3 bbmle_1.0.24
[45] carData_3.0-5 Biostrings_2.58.0 vctrs_0.4.0 ggalt_0.4.0
[49] extrafontdb_1.0 DelayedMatrixStats_1.12.3 stringr_1.4.0 beachmat_2.6.4
[53] lifecycle_1.0.1 irlba_2.3.5 gtools_3.9.2 rstatix_0.7.0
[57] XML_3.99-0.9 zlibbioc_1.36.0 MASS_7.3-56 scales_1.1.1
[61] vroom_1.5.7 hms_1.1.1 proj4_1.0-11 curl_4.3.2
[65] memoise_2.0.1 ggrastr_1.0.1 emdbook_1.3.12 bdsmatrix_1.3-4
[69] stringi_1.7.6 RSQLite_2.2.12 genefilter_1.72.1 caTools_1.18.2
[73] zip_2.2.0 BiocParallel_1.24.1 rlang_1.0.2 pkgconfig_2.0.3
[77] bitops_1.0-7 lattice_0.20-45 purrr_0.3.4 labeling_0.4.2
[81] cowplot_1.1.1 bit_4.0.4 tidyselect_1.1.2 plyr_1.8.7
[85] R6_2.5.1 generics_0.1.2 DelayedArray_0.16.3 DBI_1.1.2
[89] pillar_1.7.0 withr_2.5.0 survival_3.3-1 abind_1.4-5
[93] RCurl_1.98-1.6 ash_1.0-15 tibble_3.1.6 crayon_1.5.1
[97] car_3.0-12 KernSmooth_2.23-20 utf8_1.2.2 BiocFileCache_1.14.0
[101] tzdb_0.3.0 progress_1.2.2 locfit_1.5-9.4 grid_4.0.4
[105] blob_1.2.3 digest_0.6.29 xtable_1.8-4 numDeriv_2016.8-1.1
[109] munsell_0.5.0 beeswarm_0.4.0 vipor_0.4.5