Hello everyone,
I performed a DeSeq2 analysis and after I performed the lfcshrinkage command, I obtained very small log2fold changes for some of my results. As a consequence, my enhanced volcano plot looks very weird.
For explaining my approach: I'm using TCGA-ovarian HT-Seq counts downloaded from the GDC Bioportal, Because there is a high amount of genes with 0s, I set the
keep <- rowSums(counts(dds)) >= 1000 to a 1000 as seen and then continued with my analysis. After performing lfcshrinkage I obtained the following volcano plot.
Now I'm trying to figure out if something is wrong with my data or what is the reason for this weird looking volcano plot.
Has anybody obtained similar results like this?
Thank you very much for any help

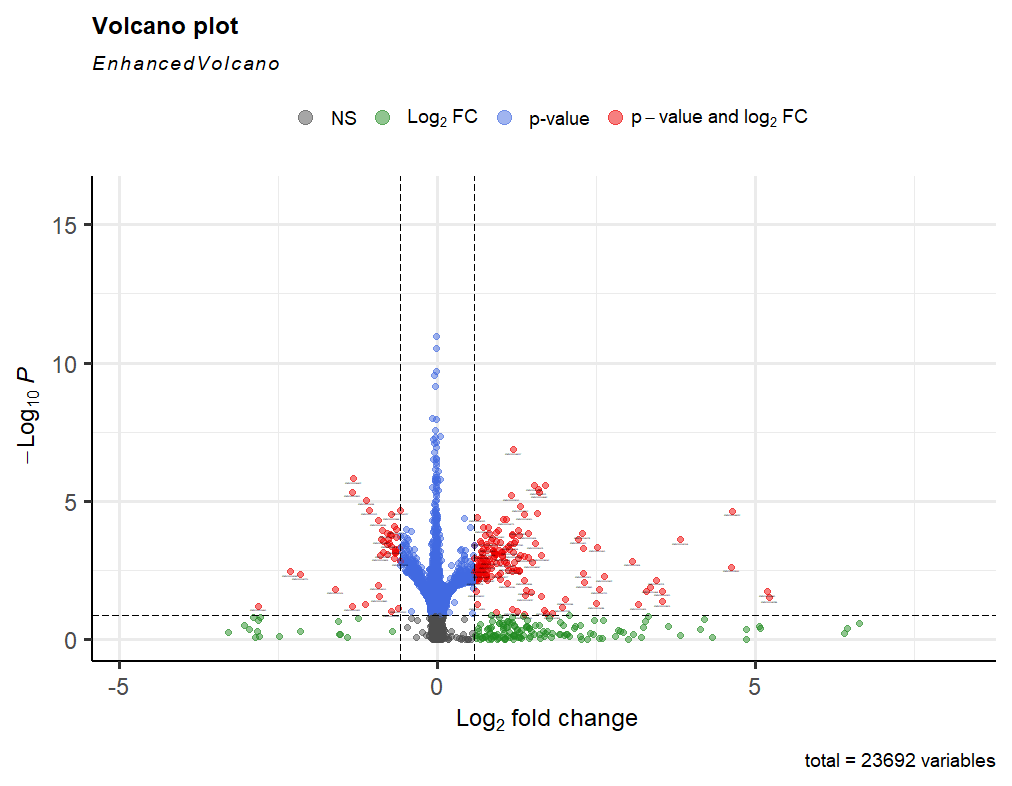
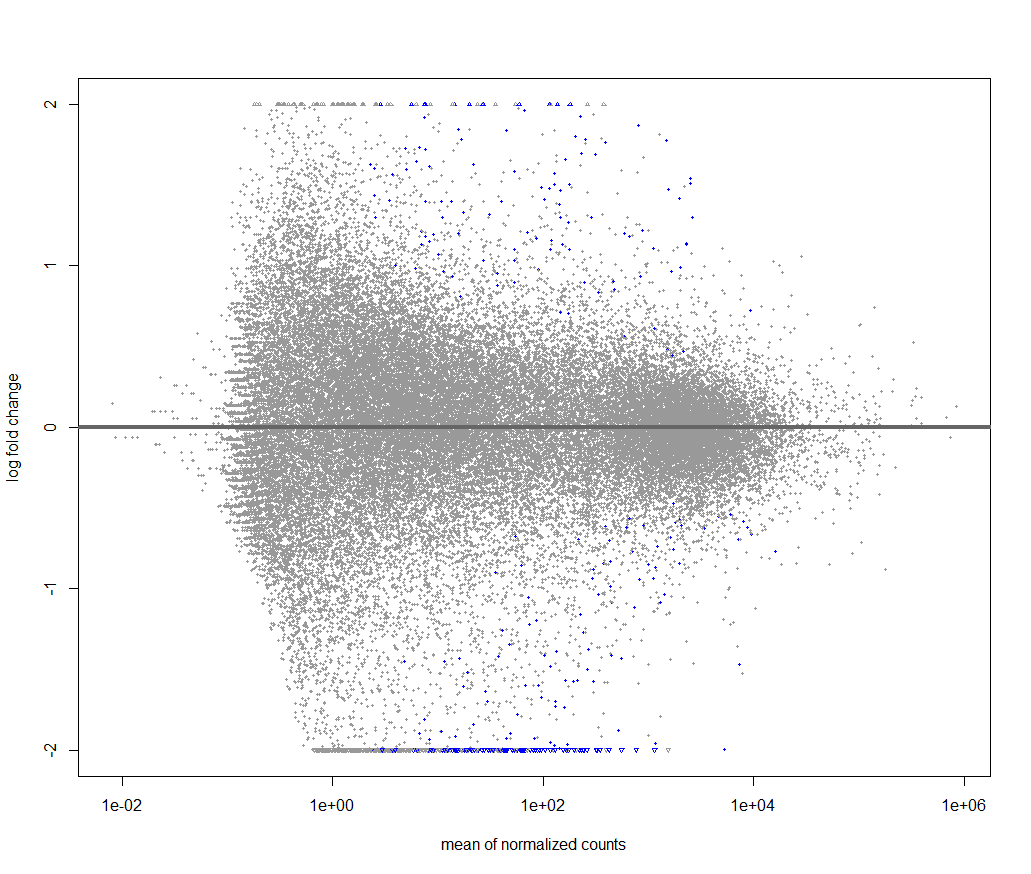
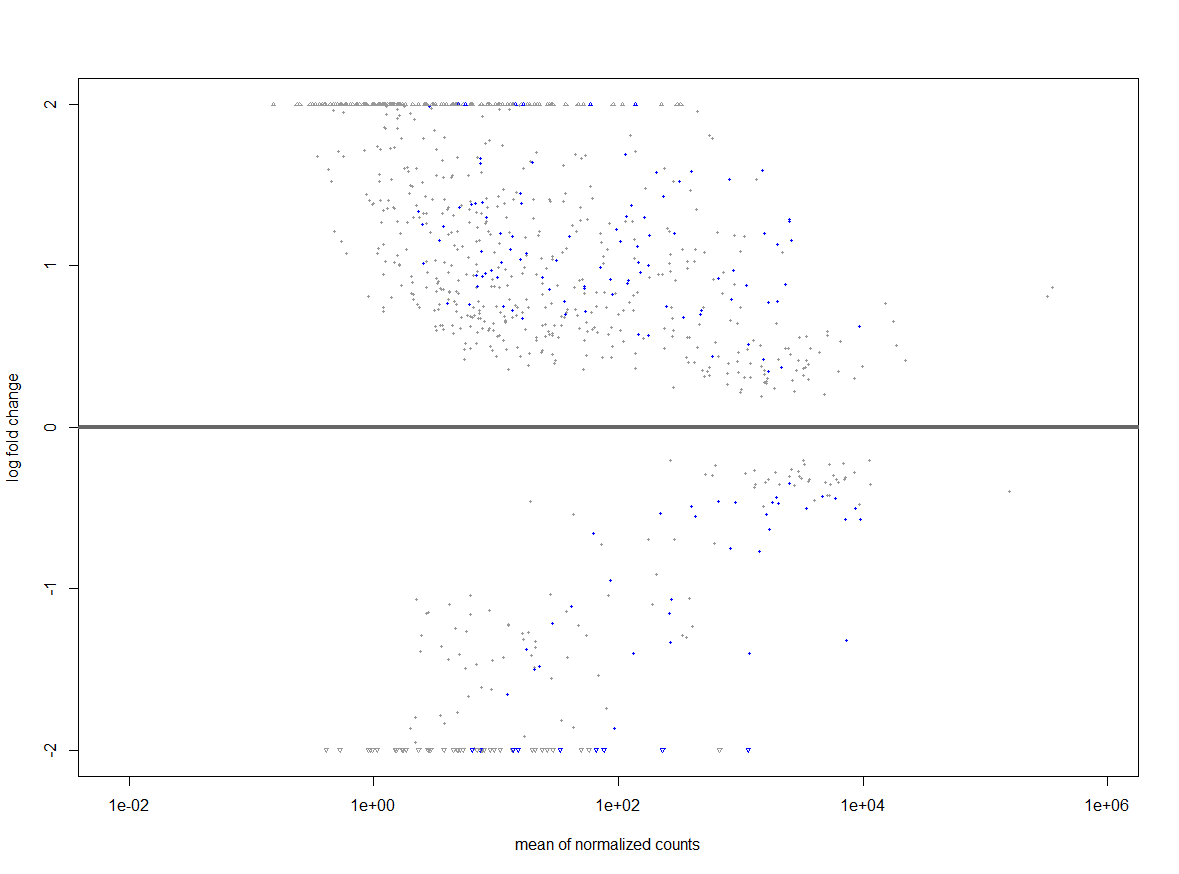

You appear to be throwing out all genes with less than 1000 total reads. Is there a reason you're setting that threshold so high? I imagine this could introduce some peculiarities in your data set. You might reduce this threshold (the vignette sets it to 10!) and see how that changes your overall results.
Thank you for the quick reply. I set the default so high, to avoid the genes with so many 0s. That's the plot with the default setting (>10).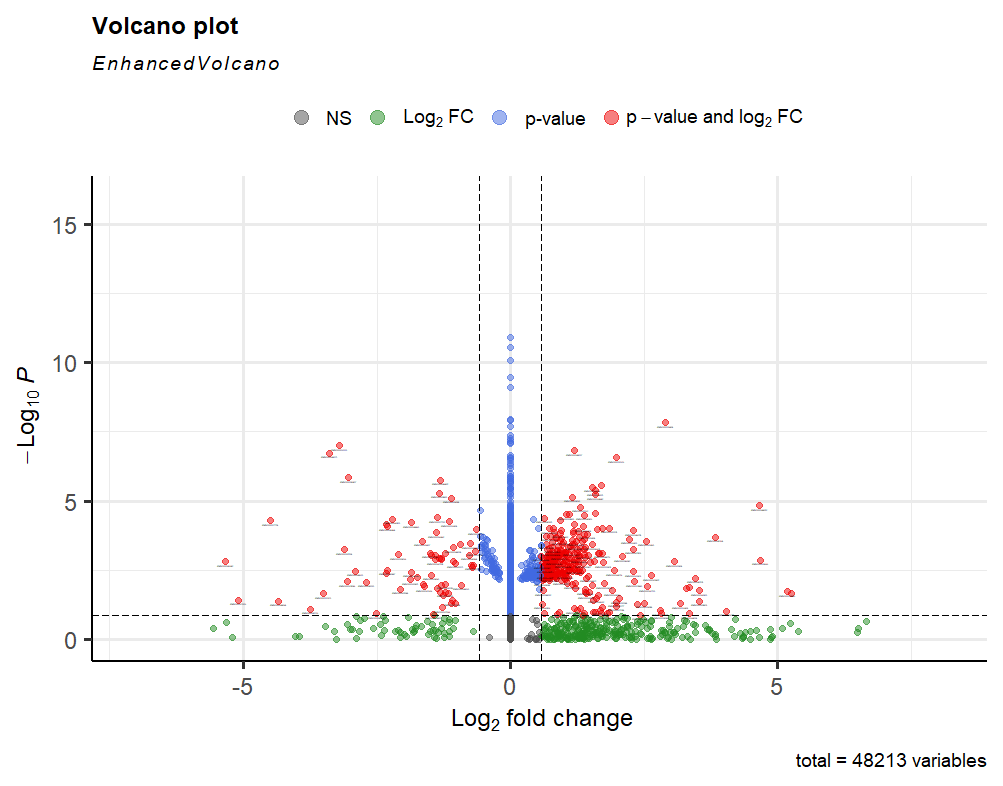
It might be a good idea to explore your data to determine why you have so many zero count genes. Do these tend to occur in particular samples more often than others?
You could also try the alternative filtering method mentioned in the vignette where you set a requirement for each gene that you must have a certain number (X) of samples with read counts greater than a threshold: