Hi everyone,
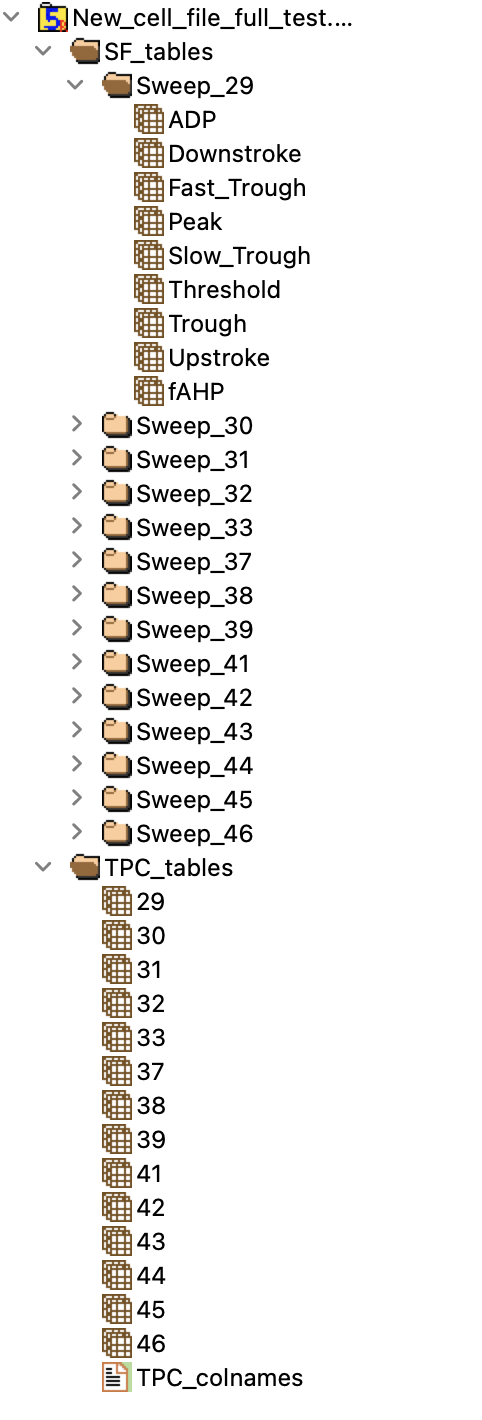
I am currently trying to read an H5 file in Rstudio using the rhdf5 package (version 2.38.1) The file is organized as shown in this image:
Here's my problem: When I try to 1)load the full TPC_tables group in a variable or 2) read TPC tables which are quite large (99999 x 5), I do not have any issue as shown in this code block:
> TPC_group=h5read(file="/Users/julienballbe/Downloads/New_cell_file_full_test.h5",name='/TPC_tables')
> head(t(TPC_group$'29'))
[,1] [,2] [,3] [,4] [,5]
[1,] 0.52002 -69.81251 0 -0.04637020 0.007849086
[2,] 0.52004 -69.81251 0 -0.04497140 0.069939827
[3,] 0.52006 -69.81251 0 -0.04270432 0.113353990
[4,] 0.52008 -69.78125 0 -0.03992725 0.138853552
[5,] 0.52010 -69.75000 0 -0.03693670 0.149527574
[6,] 0.52012 -69.75000 0 -0.03394280 0.149694873
> TPC=h5read(file="/Users/julienballbe/Downloads/New_cell_file_full_test.h5",name='/TPC_tables/29')
> head(t(TPC))
[,1] [,2] [,3] [,4] [,5]
[1,] 0.52002 -69.81251 0 -0.04637020 0.007849086
[2,] 0.52004 -69.81251 0 -0.04497140 0.069939827
[3,] 0.52006 -69.81251 0 -0.04270432 0.113353990
[4,] 0.52008 -69.78125 0 -0.03992725 0.138853552
[5,] 0.52010 -69.75000 0 -0.03693670 0.149527574
[6,] 0.52012 -69.75000 0 -0.03394280 0.149694873
However, when I try to load SF_tables group, or sub groups (like "Sweep_29"), which are only composed of at most 20-elements array into a variable, I get the following error
> SF_group=h5read(file="/Users/julienballbe/Downloads/New_cell_file_full_test.h5",name='/SF_tables')
Error in H5Dread(h5dataset = h5dataset, h5spaceFile = h5spaceFile, h5spaceMem = h5spaceMem, :
Not enough memory to read data! Try to read a subset of data by specifying the index or count parameter.
Error: Error in h5checktype(). H5Identifier not valid.
> SF=h5read(file="/Users/julienballbe/Downloads/New_cell_file_full_test.h5",name='/SF_tables/Sweep_29')
Error in H5Dread(h5dataset = h5dataset, h5spaceFile = h5spaceFile, h5spaceMem = h5spaceMem, :
Not enough memory to read data! Try to read a subset of data by specifying the index or count parameter.
I suppose, but I am not certain, that the Error comes from the fact that some of these array may be empty, because if I delete those empty arrays, the error does not appear, but why does it return 'Not enough memory to read data'?
Thank you for any help you can give me!
Best Julien


Thanks for the report. I think the error message is probably misleading. IIRC that gets triggered when memory allocation fails, and the message assumes that was because your requested something too large. However there are other reasons it could fail.
It seems likely the empty datasets you mentioned are tripping a bug in the code somewhere. I wonder if it requests 0 bytes of memory and then treats that like a failure.
I won't get round to looking at this before January, but I've created an issue on GitHub to remind me to look at this https://github.com/grimbough/rhdf5/issues/118
Hi julien.ballbe
Is it possible for you to run the
h5ls()command on your file, so I can really see what dimensions the offending datasets are. I can't reproduce this error when creating "empty" datasets myself.Something like
h5ls("/Users/julienballbe/Downloads/New_cell_file_full_test.h5")should be sufficient.If the output is huge, feel free to only include the stuff relevant to
Sweep_29.