Entering edit mode
I have run DeSeq2 in 2 different devices using the same count data and metadata tables, however when running the summary and results I get different values.
I made sure the files are read correctly, so decimals are separated by (.) in both devices and also after round the values I get the same count table values in both,
# sampleTable #
csvfile <- "metadata.csv"
sampleTable <- read.csv(csvfile, row.names=1, sep=",")
sampleTable$Stage <- as.factor( sampleTable$Stage )
summary(sampleTable)
# data matrix #
count_data_file <- "tpm.csv"
countdata <- read.csv(count_data_file, row.names=1, header=T, sep="\t", dec ="."))
summary(countdata)
# construction of DESeqDataSet #
ddsMat <- DESeqDataSetFromMatrix( countData=round(countdata), colData=sampleTable, design= ~ Stage )
nrow(ddsMat)
# removal of not or low expressed genes #
dds <- ddsMat[ rowSums(counts(ddsMat)) > 1, ]
nrow(dds)
# differential expression analysis #
dds <- DESeq(dds)
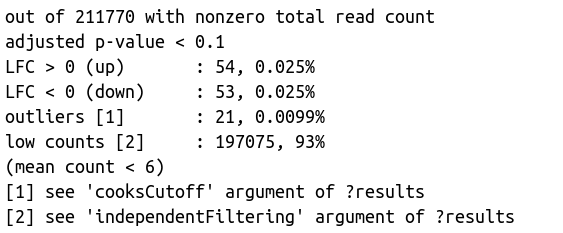
res <- results(dds)
summary(res)
sessionInfo( )