Entering edit mode
Hello,
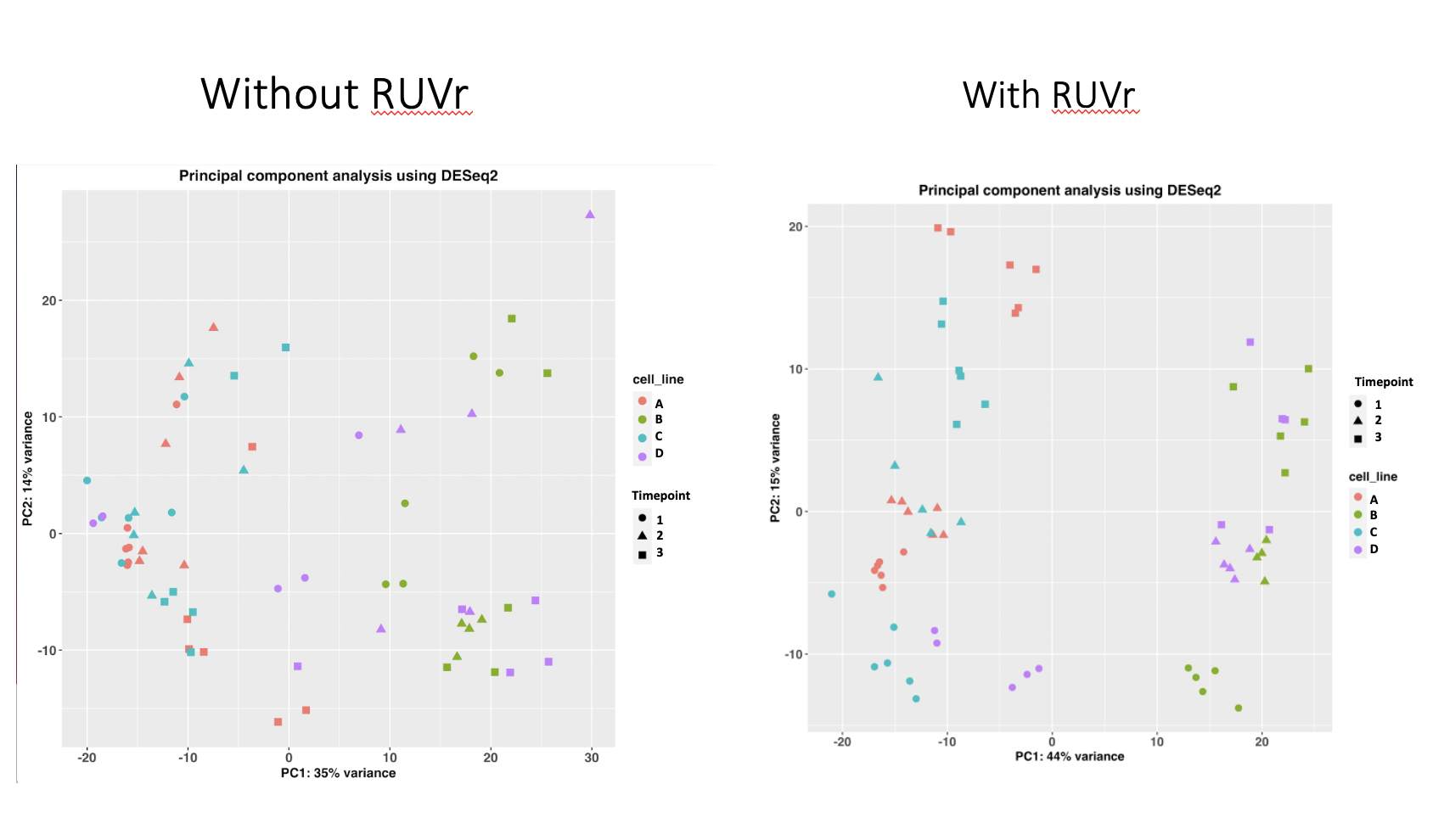
I am working on a dataset with an n=96 (four genotypes, three treatment timepoints). I am using RUVr to correct for factors of unwanted variation using k=4. After running RUVr, my PCA plot shows a much better clustering based off of genotype and timepoint. However, the number of upregulated DEGs I get (padj<0.01 and LFC>0) almost doubles for each comparison I make. Is RUVr overcorrecting the data?


Dear Michael, Thank you so much for your answer! As a follow-up, with RUVr I currently have over 4,000 upregulated DEGs (padj>0.05 and LFC>0). I would like to be more stringent so I can go with a greater LFC threshold. However, when I run a confirmation experiment (qPCR), even genes with a low LFC (0.2) are showing up as upregulated. How do I determine a threshold for a biologically relevant signal, without loosing any important information?
So you can confirm that even low LFC (by RNA-seq) genes via qPCR. This just means that you are well powered to find many differences in your system.
What's the concern? You can rank these genes by their LFC.
You need to define biological relevance with respect to an end point. Do you mean, which genes are related to a particular phenotype?
Yes sorry to clarify, I’m interested in comparing each genotype’s response to the treated timepoints. I’ve ranked my genes using LFCshrink, how do I know where to set the LFC threshold when there are so many genes showing up due to treatment? My concern is leaving out the genes with even a low LFC.
Then don't leave them out, they are just genes with lower LFC, but still not = 0
Thanks Michael! This has really helped me put together my analysis!