I got an RNA-Seq project where the data looks like as in the PCA plot below - 4 biological replicates of 4 conditions, and there are 4 batches with each batch consisting of 1 sample from each condition. In essence the variation between biological replicates is larger than the variation in the tested conditions most likely due to batch effects. Likely the wet-lab has processed each replicate of all conditions at different days.
I was thinking to include a batch into the design formulae, but on the other hand (as far as I understand) it will "remove" the variation between biological replicates, so I was wondering if that analysis would make sense?
Any advice is much appreciated
DISCLOSURE:
Cross-posted in Biostars a week ago https://www.biostars.org/p/9562841/#9563062

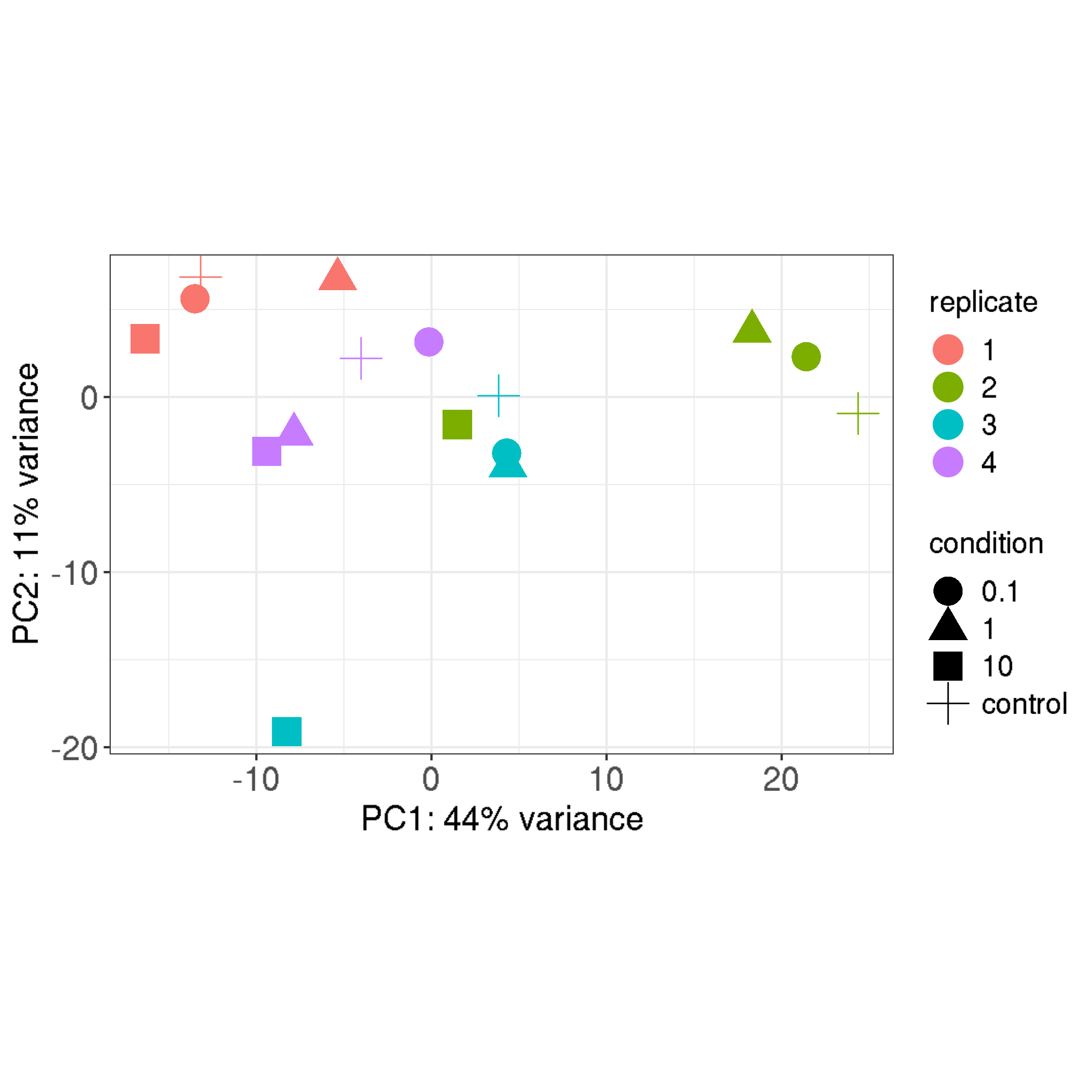

Many thanks for clarifying Mike! Just one last doubt regarding the fact that this design will control for the difference between biological replicates, does it somehow affect the final interpretation of the results? Thank you once again