Hi all,
Would you please suggest what wrong in this case? I pretty much run the same code as before. Thank you so much.
result <- dba(sampleSheet=samples)
result <- dba.blacklist(result)
result <- dba.count(result, bParallel=FALSE)
result <- dba.count(result)
result <- dba.normalize(result)
result <- dba.contrast(result, minMembers=2)
result <- dba.analyze(result)
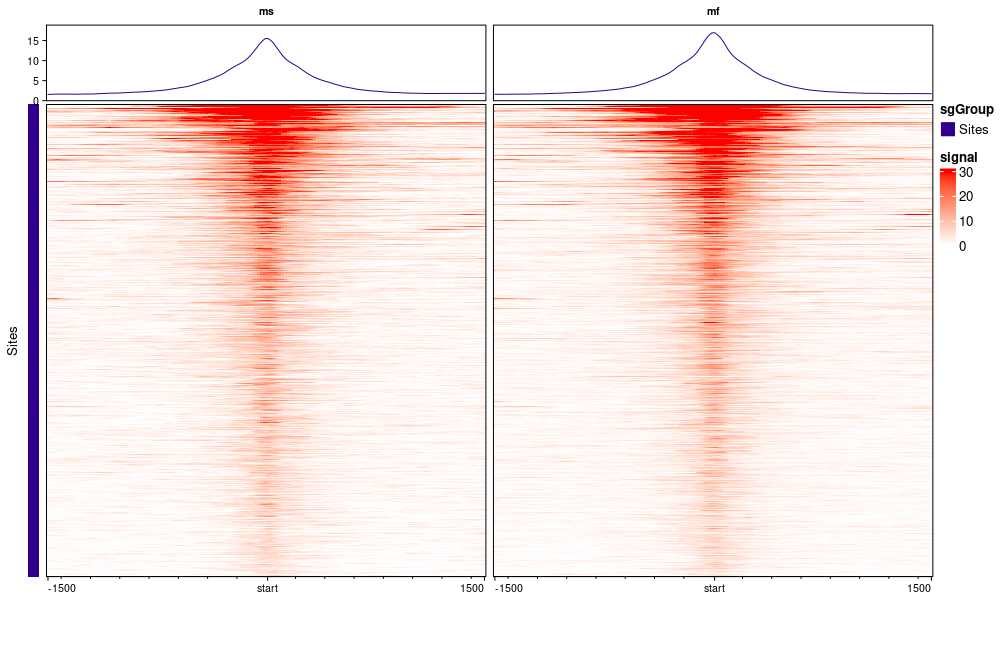
result <- dba.plotProfile(result)
Generating report-based DBA object... Error: No valid contrasts/methods specified.
And I don't know why my plot from dba.plotProfile() haven't analyzed. Would you help with this? There are two more conditions but to make it simpler, I only keep ms and mf in the samplesheet file.
dba.show(result, bContrasts=TRUE)
Factor Group Samples Group2 Samples2 DB.DESeq2
1 Condition diseased 2 control 2 0
I don't know why the value of DB.DESeq2 in this case was 0 which maybe the reason of the error. If I replace ms by other condition, I am no longer has the error.