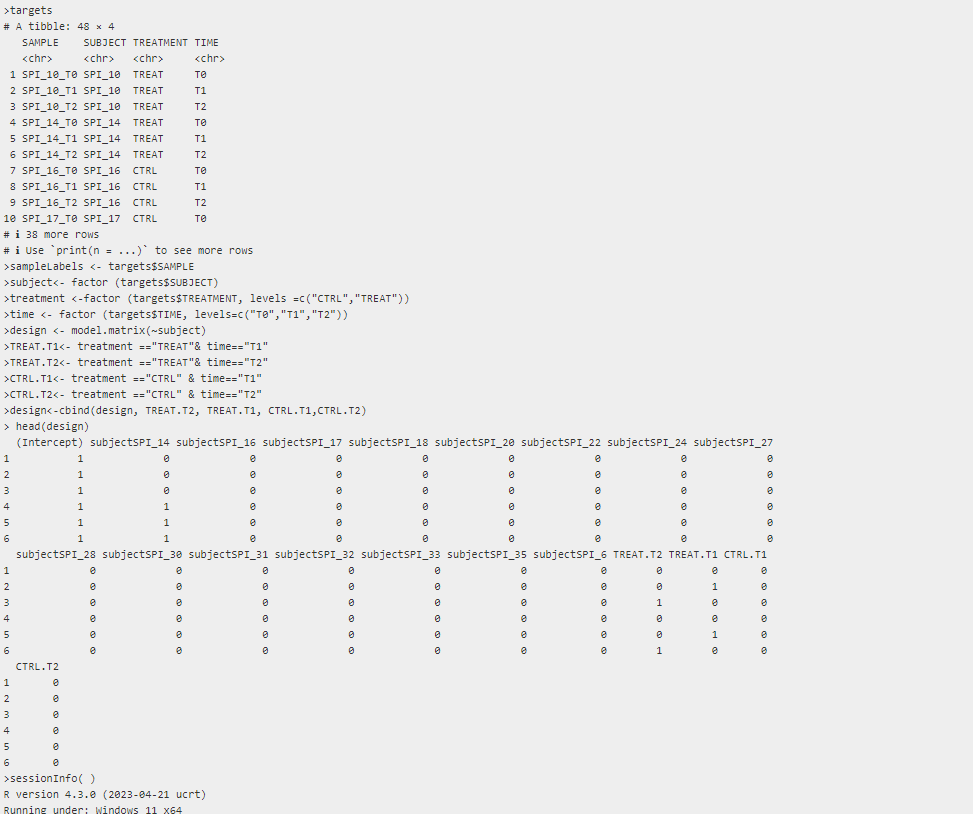
Hello, I'm trying to analyze a dataset in which there are repeated measures using edgeR. I have 48 RNA samples, collected from 16 subjects (3 time points: T0, T1, T2): 8 subjects belong to the group placebo, and the other 8 to the group treatment. So we can consider T0 as an internal control and the placebo group as an external one. I have read the 3.5 section of edgeR manual and taking that example in consideration I design a matrix as followed. If I use the coefficient "TREAT.T1" in the differential expression analysis (LRT) I will obtained the genes that are differentially expressed comparing "TREAT.T1" vs subject (TREAT.T0), is it right? and the same for "TREAT.T2", "CTRL.T1", "CTRL.T2"... If I set "subject" as an intercept I will compare each T1/T2 condition with the corrisponding T0 that in my case is not explicitly defined because it corresponds to the "subject". So not only the first subject (SPI_10) will become the intercept, but all of them according to the treatment, is it right? Thank you in advance for your attention Greta
EdgeR and repeated measures: how to design a correct matrix having three time points
0
Entering edit mode
Similar Posts
Loading Similar Posts
Traffic: 1336 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thank you for your help