Entering edit mode
Hello all,
I am trying to use gDNAx to detect gDNA contamination and filter them from my RNA-Seq data. I have tried running the package according to the vignette, and i have questions regarding the diagnostic output.
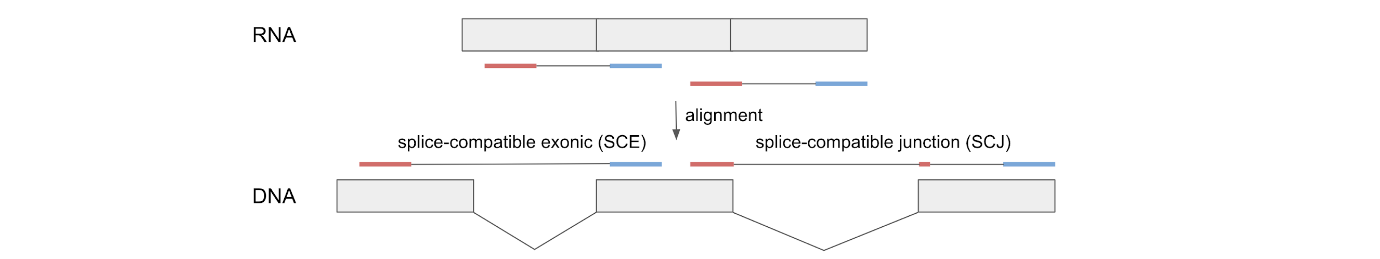
- SCJ is described as splice alignments overlapping a transcript in a "splice compatible" way. As with SCE being alignments without a splice site, but overlap a transcript in a "splice compatible" way. What does it mean by being "splice compatible", and how is this determined from the data?
- Aside from IGC (Intergenic), INT (intronic), SCJ and SCE. What are the other origins of alignments: SCC, IGCFLM, SCJFLM, SCEFLM and INTFLM?
gDNAx looks like a very promising tool, i would appreciate any details you can share.
Many thanks.