Dear bioconductor community,
I have recently analyzed two EPIC methylation array datasets using the same pipeline in R. Both of the datasets have the same variables and the comparison was PET Amyloid negative vs PET amyloid positive samples in order to find differentially methylated CpG sites. Both of the datasets are from EPIC array, but different batch(batch 0 and batch 1).
I've already tried to correct the batch effect using Harman batch correction
shifted_betas <- shiftBetas(betas=getBeta(mSetFunnormFlt), shiftBy=1e-4)
#mSetFunnormFlt is GenomicRatioSet which is the result of probe QC and Funnorm normalization
shifted_ms <- beta2m(shifted_betas)
plot(density(shifted_ms, 0.05), main="Shifted M-values, shiftBy = 1e-4",
cex.main=0.7)
shifted_ms
methHarman <- harman(shifted_ms, expt=targets_pr$Sample_Group,
batch=targets_pr$batch, limit=0.65)
#Sample_Group was divided into P and N (PET amyloid positive and Negative)
ms_hm <- reconstructData(methHarman)
fit <- lmFit(ms_hm, design)
# design matrix was created by using the code below
# design <- model.matrix(~0+Sample_Group+Age+Sex+Center+Smoking+Bcell+CD4T+CD8T+Mono+NK+Neu, data=targets_pr_0)
After batch correction, I could not get any significant differentially methylated CpG site (PET Amyloid positive vs PET Amyloid negative)
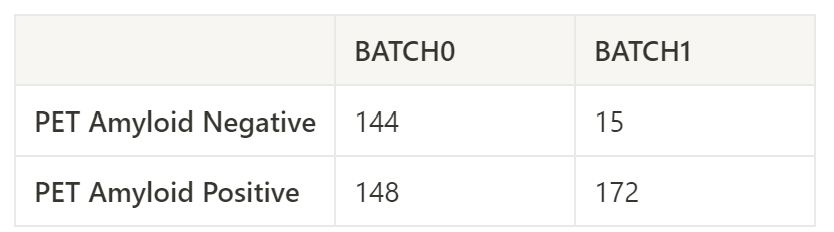
When I did methylation analysis with same pipeline separately (batch0 and batch1), I found 2,756 significant DMCs from batch 0 but 0 significant DMCs from batch 1.
I thought that case-control unbalance may cause something wrong.
My question is whether it is possible and applicable in someway to correct the batch effect in unbalanced case-control dataset?
Thank you in advance.
Best,
Yujin Kim.


Thank you for your response Gordon :)
I've already run the code below, just add batch to the limma linear model
It results in 1 significant DMC.
What do you think about using removeBatchEffect in limma in my pipeline?
Is it okay to apply removeBatchEffect before a limma analysis(limma linear model)?
Sorry for basic question, I am new to limma analysis.
Are there any useful documents I can study related to limma?
I already read the limma user's guide but there was no explanation provided regarding the batch effect.(removeBatchEffect)
I agree with your words
I need to concern that I should use the analysis of batch 0 only.
Thanks a lot Gordon!
Best,
Yujin Kim
No, you should not apply any batch correction before a limma differential analysis. The removeBatchEffect() help page that you would see by typing
?removeBatchEffectadvises you it should not be used in a differential analysis.The User's Guide and the help page for each function.
removeBatchEffect is not mentioned because it is not recommended as part of a differential analysis. Batch correction is instead done as part of the linear model, same as for any blocking variable.
PS. Just to clarify, we have seen good results from RUV normalization and background correction for large datasets. However, when the datasets is of moderate size and the batch factor is known, the correction is better done as part of the linear model.
Thank you for your kind response Gordon! :)
Have a good time at the end of the year!
Best,
Yujin Kim