I'm trying to map a genelist of NCBI Gene IDs from Scomber japonicus (Chub mackerel) to Danio rerio (Zebrafish) for further analysis with tools tailored to Danio that are not available for S japonicus.
I noticed that while Orthology.eg.db is able to convert most Scomber japonicus genes to NCBI Gene IDs, it fails to map LOC genes between species. For my analysis, I don't necessarily care that these genes aren't actually orthologs and that in both species these genes are LOC genes. I am just interested in programmatically converting my genelist of NCBI Gene IDs between the two species while capturing LOC to LOC conversions in addition to ortholog mapping. Is there a way to get around this limitation of Orthology.eg.db for LOC genes or is there another tool that I can use since they are not strictly orthologs?
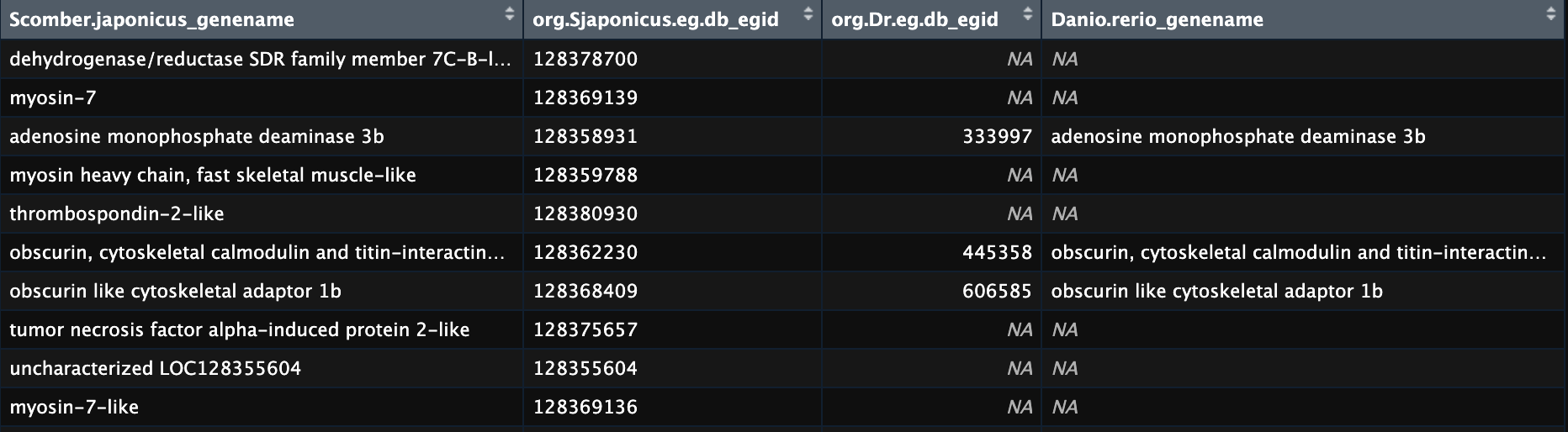
Example using a genelist of NCBI Gene IDs:
genelist <- c(
"128378700", "128369139", "128358931", "128359788", "128380930",
"128362230", "128368409", "128375657", "128355604", "128369136"
)
mappings <- AnnotationDbi::select(
Orthology.eg.db,
keys = genelist,
"Danio.rerio",
"Scomber.japonicus"
)
This is a screenshot of the results from my example with some annotation added
Thank you for taking the time to answer and sorry if this is a dumb question!