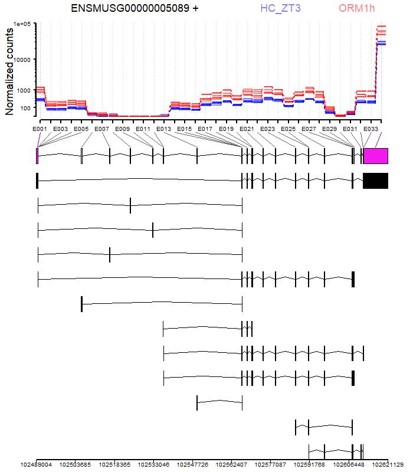
Entering edit mode
Dear, I m using DEXSeq in order to analyse DEU. I got a result table with DEXSeq() function, I got many interesting differential exon usage with padj <0.05 but for one of them I got 2 exons with padj very low but in result table I got NA values for groups values and log2foldchange as:
groupID featureID exonBaseMean dispersion stat pvalue padj HC_ZT3 ORM1h
ENSMUSG00000005089:E001 ENSMUSG00000005089 E001 685.2941 0.001725370 33.03974 9.029436e-09 1.992781e-05 NA NA
ENSMUSG00000005089:E034 ENSMUSG00000005089 E034 45712.1062 0.001449945 14.68005 1.273875e-04 4.215027e-02 NA NA
log2fold_ORM1h_HC_ZT3 genomicData.seqnames genomicData.start genomicData.end genomicData.width genomicData.strand
ENSMUSG00000005089:E001 NA 2 102489004 102489567 564 +
ENSMUSG00000005089:E034 NA 2 102612089 102621129 9041 +
countData.NC_HC_AM1 countData.NC_HC_AM2 countData.NC_HC_AM3 countData.NC_HC_AM4 countData.NC_HC_AM5 countData.NC_HC_AM6
ENSMUSG00000005089:E001 343 301 338 356 301 398
ENSMUSG00000005089:E034 24285 24536 26479 28804 25330 33677
countData.NC_ORM1h1 countData.NC_ORM1h2 countData.NC_ORM1h3 countData.NC_ORM1h4 countData.NC_ORM1h5 countData.NC_ORM1h6
ENSMUSG00000005089:E001 1014 1221 1390 1099 896 822
ENSMUSG00000005089:E034 57453 78122 96298 68502 51991 49220
transcripts
ENSMUSG00000005089:E001 ENSMUST0....
ENSMUSG00000005089:E034 ENSMUST0....
with that result, I wanted to draw a plot with:
DTU_exon.usage_NC <- plotDEXSeq( dxr.fc_NC, "ENSMUSG00000005089",
legend=TRUE,
cex.axis=1.2, #1.2
cex=1.5,
lwd=2,
color=c("#0000FF80", "#FF000080"),
expression=FALSE,
splicing=TRUE,
displayTranscripts=TRUE)
and I got:
Fit for gene/exon ENSMUSG00000005089 threw the next warning(s): the matrix is either rank-deficient or indefinite
and
> plot(DTU_exon.usage_NC)
Error in plot.window(...) : 'xlim' nécessite des valeurs finies
In addition: Warning messages:
1: In min(x) : no non-missing arguments to min; returning Inf
2: In max(x) : no non-missing arguments to max; returning -Inf
3: In min(x) : no non-missing arguments to min; returning Inf
4: In max(x) : no non-missing arguments to max; returning -Inf
Is there a solution to fix the issue? and something is strange because I could get a plot with the following code:
DTU_exon.usage_NC <- plotDEXSeq( dxr.fc_NC, "ENSMUSG00000005089",
legend=TRUE,
cex.axis=1.2, #1.2
cex=1.5,
lwd=2,
color=c("#0000FF80", "#FF000080"),
expression=FALSE,
splicing=FALSE,
displayTranscripts=TRUE,
norCounts = T)
Thanks in advance
-Jc