Entering edit mode
I am comparing the results of DESeq2 'background' normalization with library size normalization with two different spike-ins (specified using the SpikeIn column in the samplesheet). I am getting the exact same Pvalues for all three methods. I did check that the DBA$norm$DESeq2$norm.facs values are different in each case. The MA plot using normalized counts also does not seem to be consistent with the DB sites called; lots of up-regulated sites called even though the distribution is globally shifted to <0 fold change.
dba.plotMA(diffbind, contrast=1, th = fdr_cutoff)
# make DBA object
diffbind <- dba(sampleSheet = samples %>% dplyr::select(-out_pref))
# set num cores and how many reads to store in memory
diffbind$config$cores <- db_config$cores-1
diffbind$config$yieldSize <- 20000000
# get counts
diffbind$config$scanbamparam <- ScanBamParam(flag = scanBamFlag(isDuplicate=FALSE, isSecondaryAlignment=FALSE, isSupplementaryAlignment=FALSE, isPaired=TRUE, isProperPair=TRUE, isNotPassingQualityControls=FALSE, isUnmappedQuery=FALSE, hasUnmappedMate=FALSE, isMinusStrand=NA, isMateMinusStrand=NA))
# get counts
diffbind <- dba.count(diffbind, summits=DB_summits, bUseSummarizeOverlaps = TRUE, bParallel = TRUE)
print(diffbind)
# normalization
if ("Spikein" %in% colnames(samples)){
diffbind <- dba.normalize(diffbind, normalize=DBA_NORM_LIB, spikein=TRUE)
} else{
diffbind <- dba.normalize(diffbind, normalize = DBA_NORM_NATIVE, background = TRUE)
}
# print the normalization methods
dba.normalize(diffbind, bRetrieve=TRUE)
comps <- list(
list("GroupA", "GroupB")
)
for (comp in comps){
diffbind <- dba.contrast(diffbind,
group1=diffbind$masks[[ comp[[1]] ]],
group2=diffbind$masks[[ comp[[2]] ]],
name1=comp[[1]],
name2=comp[[2]])
}
dba.show(diffbind, bContrasts=TRUE)
diffbind <- dba.analyze(diffbind, bBlacklist=FALSE, bGreylist=FALSE)
dba.show(diffbind, bContrasts = TRUE)
## R version 4.2.1 (2022-06-23)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: AlmaLinux 9.0 (Emerald Puma)
##
## Matrix products: default
## BLAS: /opt/R/R-4.2.1/lib64/R/lib/libRblas.so
## LAPACK: /opt/R/R-4.2.1/lib64/R/lib/libRlapack.so
##
## locale:
## [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
## [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
## [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] enrichplot_1.18.4
## [2] readxl_1.4.2
## [3] openxlsx_4.2.5.2
## [4] ReactomePA_1.42.0
## [5] DOSE_3.24.2
## [6] clusterProfiler_4.6.2
## [7] org.Mm.eg.db_3.16.0
## [8] ChIPseeker_1.34.1
## [9] TxDb.Mmusculus.UCSC.mm10.knownGene_3.10.0
## [10] GenomicFeatures_1.50.4
## [11] AnnotationDbi_1.60.2
## [12] cowplot_1.1.1
## [13] ggplot2_3.4.2
## [14] lattice_0.21-8
## [15] gridExtra_2.3
## [16] patchwork_1.1.2
## [17] rtracklayer_1.58.0
## [18] Rsamtools_2.14.0
## [19] Biostrings_2.66.0
## [20] XVector_0.38.0
## [21] kableExtra_1.3.4
## [22] DiffBind_3.8.4
## [23] SummarizedExperiment_1.28.0
## [24] Biobase_2.58.0
## [25] MatrixGenerics_1.10.0
## [26] matrixStats_0.63.0
## [27] GenomicRanges_1.50.2
## [28] GenomeInfoDb_1.34.9
## [29] IRanges_2.32.0
## [30] S4Vectors_0.36.2
## [31] BiocGenerics_0.44.0
## [32] readr_2.1.4
## [33] stringr_1.5.0
## [34] dplyr_1.1.2
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.3
## [2] tidyselect_1.2.0
## [3] RSQLite_2.3.1
## [4] htmlwidgets_1.6.2
## [5] grid_4.2.1
## [6] BiocParallel_1.32.6
## [7] scatterpie_0.1.9
## [8] munsell_0.5.0
## [9] codetools_0.2-19
## [10] interp_1.1-4
## [11] systemPipeR_2.4.0
## [12] withr_2.5.0
## [13] colorspace_2.1-0
## [14] GOSemSim_2.24.0
## [15] filelock_1.0.2
## [16] highr_0.10
## [17] knitr_1.42
## [18] rstudioapi_0.14
## [19] labeling_0.4.2
## [20] bbmle_1.0.25
## [21] GenomeInfoDbData_1.2.9
## [22] mixsqp_0.3-48
## [23] hwriter_1.3.2.1
## [24] polyclip_1.10-4
## [25] bit64_4.0.5
## [26] farver_2.1.1
## [27] downloader_0.4
## [28] treeio_1.22.0
## [29] coda_0.19-4
## [30] vctrs_0.6.2
## [31] generics_0.1.3
## [32] gson_0.1.0
## [33] xfun_0.39
## [34] BiocFileCache_2.6.1
## [35] R6_2.5.1
## [36] apeglm_1.20.0
## [37] graphlayouts_0.8.4
## [38] invgamma_1.1
## [39] locfit_1.5-9.7
## [40] bitops_1.0-7
## [41] cachem_1.0.7
## [42] fgsea_1.24.0
## [43] gridGraphics_0.5-1
## [44] DelayedArray_0.24.0
## [45] BiocIO_1.8.0
## [46] scales_1.2.1
## [47] ggraph_2.1.0
## [48] gtable_0.3.3
## [49] tidygraph_1.2.3
## [50] rlang_1.1.3
## [51] systemfonts_1.0.4
## [52] splines_4.2.1
## [53] lazyeval_0.2.2
## [54] yaml_2.3.7
## [55] reshape2_1.4.4
## [56] qvalue_2.30.0
## [57] tools_4.2.1
## [58] ggplotify_0.1.0
## [59] gplots_3.1.3
## [60] jquerylib_0.1.4
## [61] RColorBrewer_1.1-3
## [62] Rcpp_1.0.10
## [63] plyr_1.8.8
## [64] progress_1.2.2
## [65] zlibbioc_1.44.0
## [66] purrr_1.0.1
## [67] RCurl_1.98-1.12
## [68] prettyunits_1.1.1
## [69] deldir_1.0-6
## [70] viridis_0.6.2
## [71] ashr_2.2-54
## [72] ggrepel_0.9.3
## [73] magrittr_2.0.3
## [74] data.table_1.14.8
## [75] reactome.db_1.82.0
## [76] truncnorm_1.0-9
## [77] mvtnorm_1.1-3
## [78] SQUAREM_2021.1
## [79] amap_0.8-19
## [80] xtable_1.8-4
## [81] hms_1.1.3
## [82] evaluate_0.23
## [83] HDO.db_0.99.1
## [84] XML_3.99-0.14
## [85] emdbook_1.3.12
## [86] jpeg_0.1-10
## [87] compiler_4.2.1
## [88] biomaRt_2.54.1
## [89] bdsmatrix_1.3-6
## [90] tibble_3.2.1
## [91] shadowtext_0.1.2
## [92] KernSmooth_2.23-20
## [93] crayon_1.5.2
## [94] htmltools_0.5.5
## [95] ggfun_0.0.9
## [96] tzdb_0.3.0
## [97] geneplotter_1.76.0
## [98] tidyr_1.3.0
## [99] aplot_0.1.10
## [100] DBI_1.1.3
## [101] tweenr_2.0.2
## [102] dbplyr_2.3.2
## [103] MASS_7.3-59
## [104] rappdirs_0.3.3
## [105] boot_1.3-28.1
## [106] ShortRead_1.56.1
## [107] Matrix_1.5-4
## [108] cli_3.6.1
## [109] parallel_4.2.1
## [110] igraph_1.4.2
## [111] pkgconfig_2.0.3
## [112] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
## [113] GenomicAlignments_1.34.1
## [114] numDeriv_2016.8-1.1
## [115] xml2_1.3.3
## [116] annotate_1.76.0
## [117] ggtree_3.6.2
## [118] svglite_2.1.1
## [119] bslib_0.4.2
## [120] webshot_0.5.4
## [121] rvest_1.0.3
## [122] yulab.utils_0.0.6
## [123] digest_0.6.34
## [124] graph_1.76.0
## [125] cellranger_1.1.0
## [126] rmarkdown_2.21
## [127] fastmatch_1.1-3
## [128] tidytree_0.4.2
## [129] restfulr_0.0.15
## [130] GreyListChIP_1.30.0
## [131] curl_5.0.1
## [132] graphite_1.44.0
## [133] gtools_3.9.4
## [134] rjson_0.2.21
## [135] lifecycle_1.0.3
## [136] nlme_3.1-162
## [137] jsonlite_1.8.7
## [138] viridisLite_0.4.1
## [139] limma_3.54.2
## [140] BSgenome_1.66.3
## [141] fansi_1.0.4
## [142] pillar_1.9.0
## [143] plotrix_3.8-2
## [144] KEGGREST_1.38.0
## [145] fastmap_1.1.1
## [146] httr_1.4.5
## [147] GO.db_3.16.0
## [148] glue_1.6.2
## [149] zip_2.3.0
## [150] png_0.1-8
## [151] bit_4.0.5
## [152] ggforce_0.4.1
## [153] stringi_1.7.12
## [154] sass_0.4.5
## [155] blob_1.2.4
## [156] DESeq2_1.38.3
## [157] latticeExtra_0.6-30
## [158] caTools_1.18.2
## [159] memoise_2.0.1
## [160] irlba_2.3.5.1
## [161] ape_5.7-1

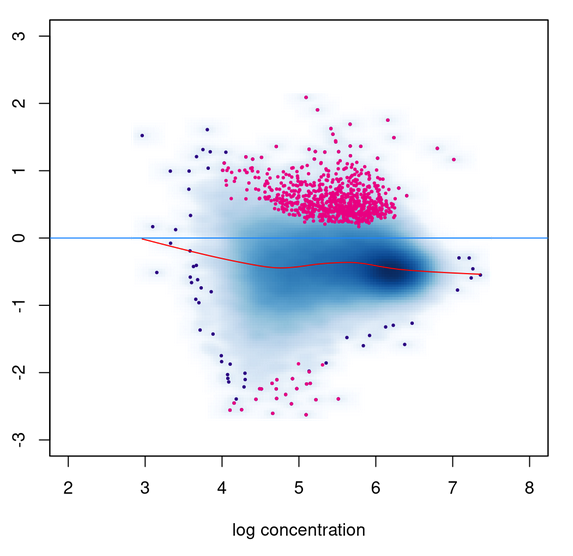

To help make it easier to reproduce this issue, I was able to reproduce similar behavior using the included tamoxifen dataset. I also realized that while the PValues are identical, the 'Conc' columns are distinct.
Packages loaded
Library size norm
Background counts RLE
Compare results
SessionInfo