Hi,
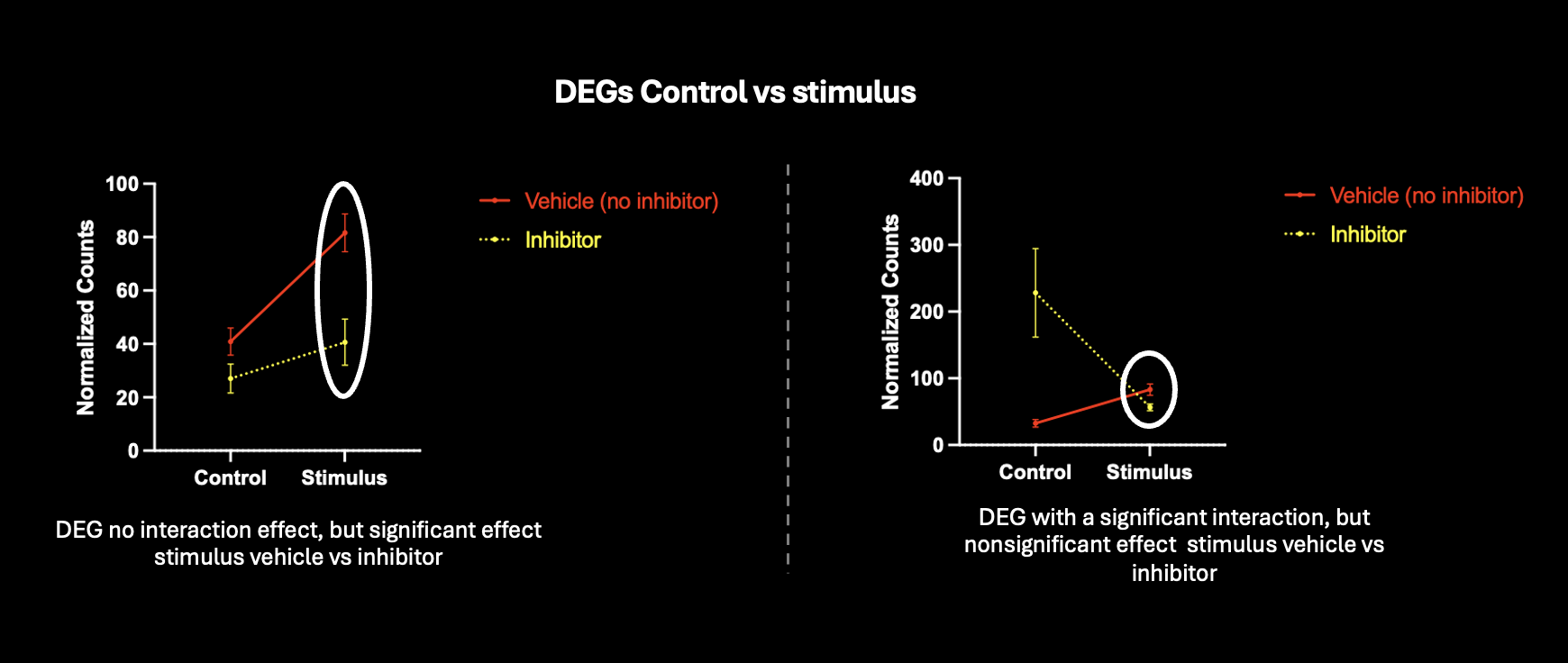
I would really appreciate input about correcting for multiple comparisons in my dataset. I've identified DEGs after exposure to a stimulus (control vs stimulus). I'm exploring whether these gene expression changes might be influenced by a protein. For this, I've inhibited the protein under control and stimulus conditions and run another differential expression comparison to see if DEGs (control vs stimulus) show an significant change after inhibitor treatment (stimulus vs stimulus + inhibitor).
Is this approach accurate, or am I introducing false positives by running two separate comparisons for differential expression ? I've tried looking at genes with an interaction, but this would take into consideration genes where the effect is different under control+inhibitor conditions. Shown are two genes that are differentially expressed control vs stimulus, and trying to run another comparison looking at stimulus vs stimulus+inhibitor
Any advice would be appreciated-thanks!