Hello All, I am new to bioinformatics this may be basic but would really appreciate any help
I have RNAseq data, sensitive and tolerant to a drug , which is treated with one drug so I have four different classes of data 1)sensitive+treated,2)sensitive+untreated , 3)tolerant+untreated and 4) tolerant +treated each having three replicates
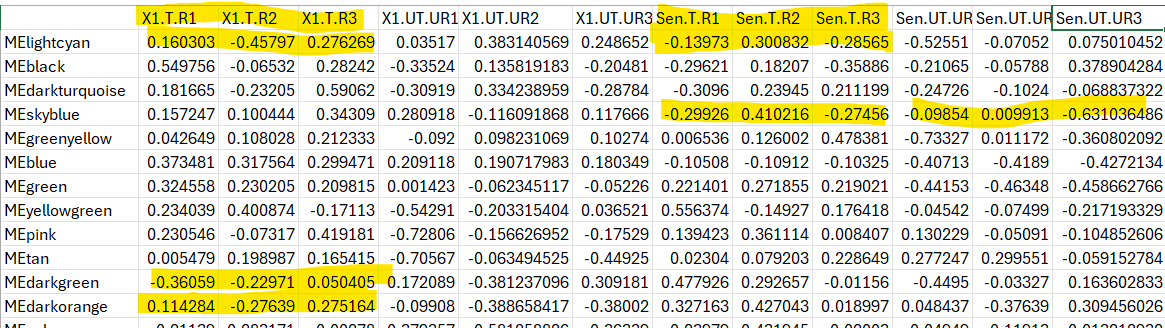
I want to perform WGCNA to find a modules that are unique in either sensitive or resistant i.e the genes that are differentially expressed in sensitive and not in resistant or vice versa. I have three replicates of each sample, so my question is how to handle the replicates because if I am considering as R1,R2,R3 they are having different module eigen values so how can I merge them to have one representative module.