Hi BioConductor Community ,
I am trying to automate the process of generating gene expression files from TSV raw series files on GEO.
Does anyone have any tips or BioConductor libraries that can generate gene expression levels from TSV files on GEO ?
I have been filtering GEO series records with the following query ("tsv"[Supplementary Files])
Thank you ,
-K


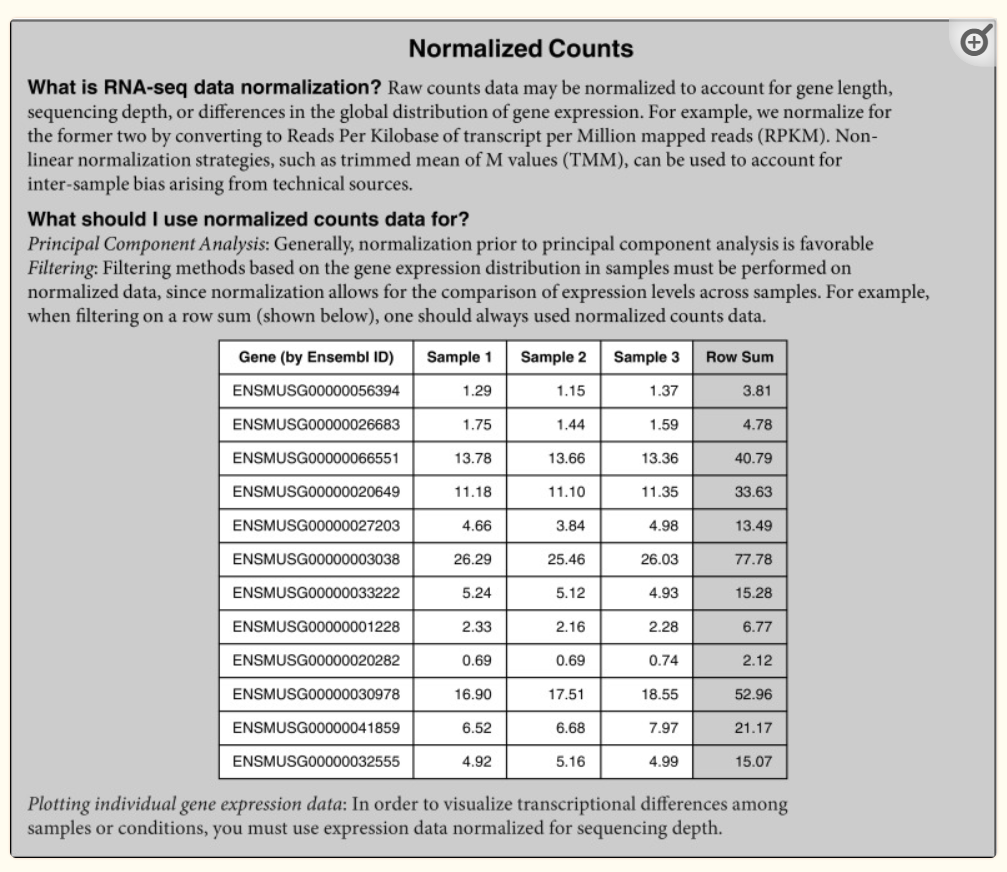
I believe most of the TSV files are RNA-Seq data , I need to automate the process for all tsv files on geo. The end product of what I need is what is shown in the table in the attached image.