Hello, I am running my GeoMx Data with Standr, and it includes the "limma" package.
I am incorporating the codes from these two websites,
https://ucdavis-bioinformatics-training.github.io/2018-June-RNA-Seq-Workshop/thursday/DE.html
and some parts are showing errors or unexpected results.
Despite the error below, I was able to run the next part of the limma DE gene analysis, but the result was 0 downreg and 0 upreg genes.
I suppose it may be due to the error below. It would be very much appreciated if someone could help.
The data structure may be a little different to scRNAseq data, since it is an spe object (SpatialExperiment class).
#Differential expression analysis with limma-voom pipeline
>library(edgeR)
>library(limma)
>HNdge <- SE2DGEList(HNspe_ruv)
>HNdesign <- model.matrix(~0 + biology + ruv_W1 + ruv_W2, data = colData(HNspe_ruv))
>mm <- model.matrix(~0 + Disease_status, data = colData(HNspe_ruv))
>y <- voom(mm, plot = F)
Error in approxfun(l, rule = 2, ties = list("ordered", mean)) : need at least two non-NA values to interpolate
fit <- lmFit(y, mm)
contr <- makeContrasts(groupI5.6 - groupC.6, levels = colnames(coef(fit)))
tmp <- contrasts.fit(fit, contr)
tmp <- eBayes(tmp)
top.table <- topTable(tmp, sort.by = "P", n = Inf)
HNcontr.matrix <- makeContrasts(
BvT = Healthy - Disease,
levels = colnames(HNdesign))
HNv <- voom(HNdge_all, HNdesign, plot = TRUE)
include your problematic code here with any corresponding output
y <- voom(mm, plot = F)
please also include the results of running the following in an R session
Error in approxfun(l, rule = 2, ties = list("ordered", mean)) : need at least two non-NA values to interpolate
sessionInfo( )
R version 4.4.1
package.version("limma") "3.60.3"
```

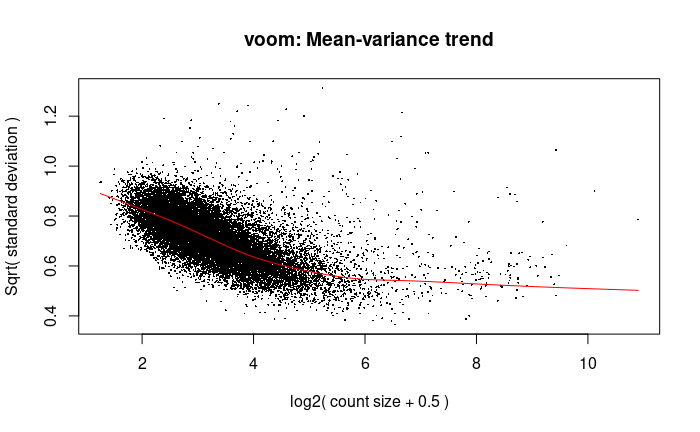

Yes I have changed it to a design matrix and it has worked. Thank you very much!
I have encountered a different problem now,
it ran through fine up until here, but then the code below is saying that the number of rows does not match. Should I compare the number of rows btw "fit" and "contr"?
How would I fix this mismatch of number of rows?
Error in contrasts.fit(fit, contrasts = contr_nevus) : Number of rows of contrast matrix must match number of coefficients in fit
You should inspect the column names of your model matrix. It's unlikely that the column names are CN and HN.
The column name is biology, and the categories in that column is CN and HN. If it only accept column names, how do I compare a certain category from a column to a certain category from the same column?
You don't compare categories within a column of a design matrix. Categorical variables should be expanded automatically by R into 'dummy variables' that contain zeros and ones only.
You will need to provide the design and contrasts matrices, as it's not clear to me what they are.