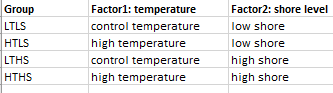
Hello, I'm working on GLM model in DEseq2 with an experimental design of 4 treatments , and each treatment has 5 replicates. I have two factors: temperature (with control and high temperature) and shore level (high shore and low shore).
I've been running a GLM model in DESeq2 using the following design:
dds <- DESeqDataSetFromMatrix(countData = counts,
colData = group,
design = ~temperature* shore_level)
In this design, the two shore levels were coded as 0 and 1, as well as the two temperature conditions. In this case, the control temperature low shore group is used as the baseline, which means the temperature main effect will only be calculated between control temp low shore and high temp low shore. However, this is actually not what I expected because, in my design, low shore is not regarded as a control group and there might be also interactions between low shore and high temp. The ideal way to measure the temperature effect would be taking the information of both shore levels into consideration.
Using a different way in coding of groups seems could retain the effect of temperature correctly, i.e. high shore encoded as 1 and low shore encoded as -1 in the glm formula. Unfortunately I didn't find a good way from the tutorial to apply this kind of design in deseq2 analysis. Could you please give me some guidance about how to set the formula in deseq2?
Any advice would be greatly appreciated. Thanks a lot!