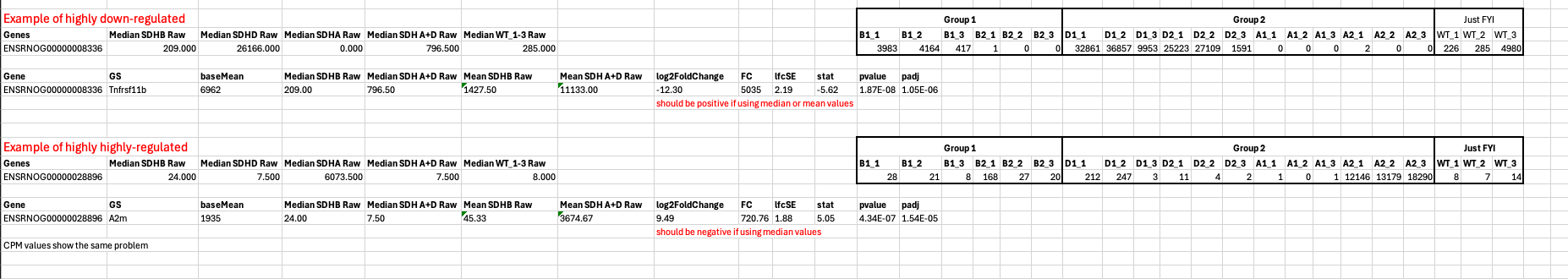
I have these two examples, where the mean and median of group vs mean and median of group 2 in two instances, I don't understand how in the first instance the log2foldchange is negative, despite the median and mean expression being lower in group 1 vs group 2.
In the second scenario the median of group 1 is higher vs group 2, whereas the mean in group 2 is higher than group 1. I don't understand how in this instance the log2foldchange is positive.
These are the raw counts, the same scenario for the same genes occurred when doing cpm transformation.
Also in both instances there is quite intra-group expression heterogeneity, how does dese2 address this?