Did you check the help page for the function enrichDO? Type ?enrichDO.
I am asking, because there it is stated that the argument ont should either be one of "HDO", "HPO" or "MPO". So "DO" (what you used) is indeed not a supported ontology!
BTW: HDO = Human Disease Ontology (link), HPO = Human Phenotype Ontology (link), MPO = Mouse Phenotype Ontology (link) )
Then it is working! Using the human example dataset (thus with the Human Disease Ontology [HDO]):
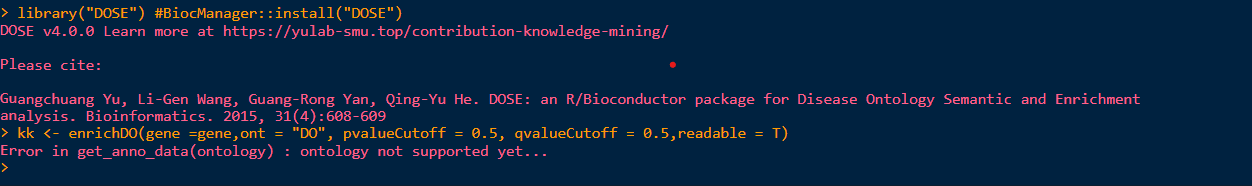
> library(DOSE)
DOSE v4.0.0 Learn more at https://yulab-smu.top/contribution-knowledge-mining/
Please cite:
Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an
R/Bioconductor package for Disease Ontology Semantic and Enrichment
analysis. Bioinformatics. 2015, 31(4):608-609
> data(geneList)
>
> gene = names(geneList)[geneList > 1]
>
> kk <- enrichDO(gene = gene,
+ ont = "HDO",
+ pvalueCutoff = 0.5,
+ qvalueCutoff = 0.5,
+ readable = T)
>
> kk
#
# over-representation test
#
#...@organism Homo sapiens
#...@ontology HDO
#...@keytype ENTREZID
#...@gene chr [1:525] "4312" "8318" "10874" "55143" "55388" "991" "6280" "2305" ...
#...pvalues adjusted by 'BH' with cutoff <0.5
#...352 enriched terms found
'data.frame': 352 obs. of 12 variables:
$ ID : chr "DOID:0060306" "DOID:934" "DOID:0080599" "DOID:11054" ...
$ Description : chr "Meier-Gorlin syndrome" "viral infectious disease" "Coronavirus infectious disease" "urinary bladder cancer" ...
$ GeneRatio : chr "8/302" "36/302" "20/302" "23/302" ...
$ BgRatio : chr "10/7865" "336/7865" "138/7865" "180/7865" ...
$ RichFactor : num 0.8 0.107 0.145 0.128 0.087 ...
$ FoldEnrichment: num 20.83 2.79 3.77 3.33 2.26 ...
$ zScore : num 12.54 6.7 6.57 6.31 5.44 ...
$ pvalue : num 1.82e-10 1.65e-08 2.58e-07 3.36e-07 1.52e-06 ...
$ p.adjust : num 1.43e-07 6.46e-06 6.59e-05 6.59e-05 1.63e-04 ...
$ qvalue : num 1.15e-07 5.20e-06 5.31e-05 5.31e-05 1.31e-04 ...
$ geneID : chr "CDC45/CDT1/MCM5/CDC6/ORC6/ORC1/GINS3/GMNN" "MMP1/S100A8/CXCL10/CXCL9/GZMB/IDO1/CCL8/CXCL3/CCL20/CXCL8/CTLA4/CCL5/MKI67/LCN2/ACE2/MX1/TREM1/LCK/C1QB/PRF1/IL"| __truncated__ "MMP1/S100A8/CXCL10/CXCL9/GZMB/CCL8/CXCL3/CXCL8/CCL5/LCN2/ACE2/C1QB/IL2RA/CCR1/STAT1/CCL2/IL2RG/CCL4/SLPI/IL7" "MMP1/S100A9/S100A8/TOP2A/RRM2/CDK1/BIRC5/EZH2/AURKA/CDC25A/PLK1/MMP9/CCNE1/MKI67/FANCA/CA9/CDC25B/TYMS/MYC/E2F1"| __truncated__ ...
$ Count : int 8 36 20 23 38 38 9 34 14 9 ...
#...Citation
Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics. 2015, 31(4):608-609
>
> as.data.frame(kk)[1:3,]
ID Description GeneRatio BgRatio
DOID:0060306 DOID:0060306 Meier-Gorlin syndrome 8/302 10/7865
DOID:934 DOID:934 viral infectious disease 36/302 336/7865
DOID:0080599 DOID:0080599 Coronavirus infectious disease 20/302 138/7865
RichFactor FoldEnrichment zScore pvalue p.adjust
DOID:0060306 0.8000000 20.834437 12.540792 1.816539e-10 1.425983e-07
DOID:934 0.1071429 2.790326 6.702107 1.645690e-08 6.459332e-06
DOID:0080599 0.1449275 3.774355 6.570134 2.583957e-07 6.594132e-05
qvalue
DOID:0060306 1.147288e-07
DOID:934 5.196914e-06
DOID:0080599 5.305370e-05
geneID
DOID:0060306 CDC45/CDT1/MCM5/CDC6/ORC6/ORC1/GINS3/GMNN
DOID:934 MMP1/S100A8/CXCL10/CXCL9/GZMB/IDO1/CCL8/CXCL3/CCL20/CXCL8/CTLA4/CCL5/MKI67/LCN2/ACE2/MX1/TREM1/LCK/C1QB/PRF1/IL2RA/CA9/CCR1/STAT1/TLR8/CCL2/PLA2G2A/MYC/IL2RG/RSAD2/CCL4/SLPI/CD163/CDKN2A/CEBPB/IL7
DOID:0080599 MMP1/S100A8/CXCL10/CXCL9/GZMB/CCL8/CXCL3/CXCL8/CCL5/LCN2/ACE2/C1QB/IL2RA/CCR1/STAT1/CCL2/IL2RG/CCL4/SLPI/IL7
Count
DOID:0060306 8
DOID:934 36
DOID:0080599 20
>
>
>


thank u so much ,god bless u,my dear friend.